For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVTTREIRLALVDDHAILRQGLRSLLEREDDLVVVGEASSEAEAEAMVAAVEPDVVLLDLKLSAGSDFEG LSLCAKLSAAHPDLGLLVLTTFLDEDLVVRAVHAGARGYVVKDVDTTELVRAIRAISSGDSAFDSRSAAA VVRSLSGRTEPREQLTDREIEVLRLLAAGLSNNKIGEKLFISATTAKFHVSNIMRKLDVSRRAEAVYAAS KRGLI*
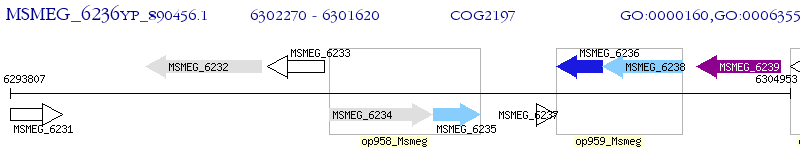
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6236 | - | - | 100% (216) | two-component system, regulatory protein |
| M. smegmatis MC2 155 | MSMEG_5244 | - | 5e-39 | 40.87% (208) | two component transcriptional regulatory protein devr |
| M. smegmatis MC2 155 | MSMEG_3944 | - | 7e-38 | 38.46% (208) | two component transcriptional regulatory protein devr |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3157c | devR | 1e-40 | 41.83% (208) | two component transcriptional regulatory protein DevR |
| M. gilvum PYR-GCK | Mflv_3847 | - | 8e-41 | 42.79% (208) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3133c | devR | 1e-40 | 41.83% (208) | two component transcriptional regulatory protein DevR |
| M. leprae Br4923 | MLBr_01286 | - | 8e-12 | 31.51% (146) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_3891c | - | 6e-42 | 41.35% (208) | LuxR family transcriptional regulator |
| M. marinum M | MMAR_3480 | - | 1e-39 | 40.48% (210) | two-component transcriptional regulatory protein |
| M. avium 104 | MAV_4109 | - | 5e-42 | 41.35% (208) | two component transcriptional regulatory protein devr |
| M. thermoresistible (build 8) | TH_2159 | - | 2e-38 | 39.90% (208) | TWO COMPONENT TRANSCRIPTIONAL REGULATORY PROTEIN DEVR (PROBABLY |
| M. ulcerans Agy99 | MUL_2423 | devR | 7e-40 | 42.03% (207) | two component transcriptional regulatory protein DevR |
| M. vanbaalenii PYR-1 | Mvan_5474 | - | 5e-99 | 88.15% (211) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6236|M.smegmatis_MC2_155 ------MTVTTREIRLALVDDHAILRQGLRSLLEREDDLVVVGEASSEAE
Mvan_5474|M.vanbaalenii_PYR-1 ----------MKPIRLVLVDDHAILRQGLRSVLERADDLIVVGEAASESE
Mb3157c|M.bovis_AF2122/97 ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
Rv3133c|M.tuberculosis_H37Rv ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
MUL_2423|M.ulcerans_Agy99 ------------MVTVFLVDDHEVVRRGLIDLLDADPQLDVVGEAGSVSE
Mflv_3847|M.gilvum_PYR-GCK ------------MVKVFLVDDHEVVRRGLIDLLGSDPELDVVGEAGSVSE
TH_2159|M.thermoresistible__bu ------------MVSVFLVDDHEVVRRGLIDLLDSDPDLEVVGEAGTVAQ
MMAR_3480|M.marinum_M ------------MVKVFLVDDHEVVRRGLVDLLGSDPDLEIIGEAGSVSE
MAV_4109|M.avium_104 ------------MVKVFLVDDHEVVRRGLCDLLSSDPDLRIVGEAGTVAE
MAB_3891c|M.abscessus_ATCC_199 ------------MITVFLVDDHEVVRRGLADLLEEEADFSVIGQASSVAE
MLBr_01286|M.leprae_Br4923 MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLR-DEGYDIVGEAADGQE
: :.:*. ::* .* .:* ::*:*. :
MSMEG_6236|M.smegmatis_MC2_155 AEAMVAAVEPDVVLLDLKLSAGSDFEGLSLCAKLSAAHPDLGLLVLTTFL
Mvan_5474|M.vanbaalenii_PYR-1 AVAVVGATSPDVVLLDLKLSAGSEFEGLTLCAKLSAAHPKLGLLVLTTFL
Mb3157c|M.bovis_AF2122/97 AMARVPAARPDVAVLDVRLPDG---NGIELCRDLLSRMPDLRCLILTSYT
Rv3133c|M.tuberculosis_H37Rv AMARVPAARPDVAVLDVRLPDG---NGIELCRDLLSRMPDLRCLILTSYT
MUL_2423|M.ulcerans_Agy99 AMARIPAANPDVAVLDVRLPDG---DGIELCRDLLSRMPNLRCLILTSYT
Mflv_3847|M.gilvum_PYR-GCK ALTRVPTAQPDVAVLDVRLPDG---NGIELCRELLSLMPDLRCLMLTSFT
TH_2159|M.thermoresistible__bu AMARIPAAQPDVAVLDVRLPDG---NGVELCRDLLARMPDLRCLMLTSYT
MMAR_3480|M.marinum_M AMARIPALQPDVAVLDVRLPDG---NGIELCRDLLSDLPELRCLMLTSFT
MAV_4109|M.avium_104 AKARIPAARPDVAVLDVRLPDG---NGIELCRDLLSEHPDLRCLMLTSFT
MAB_3891c|M.abscessus_ATCC_199 AMARIPALQPDIAVLDIRLPDG---NGVELCRELRSKLPNLNCLMLTSFT
MLBr_01286|M.leprae_Br4923 AVELAERHKPDLVIMDVKMPRR---DGIDAASEIASKRIAP-IVVLTAFS
* **:.::*:::. :*: . .: : ::**::
MSMEG_6236|M.smegmatis_MC2_155 DEDLVVRAVHAGARGYVVKDVDTTELVRAIRAISSGDSAFDSRSAAAVVR
Mvan_5474|M.vanbaalenii_PYR-1 DEDLVVRAVHAGARGYVVKDVDTTELVRAIRAISAGDSAFDSRSAAAVVR
Mb3157c|M.bovis_AF2122/97 SDEAMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMA
Rv3133c|M.tuberculosis_H37Rv SDEAMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMA
MUL_2423|M.ulcerans_Agy99 SDEAMLDAILAGASGYVVKDIKGMELARAIKEVGAGRSLLDNRAAAALMS
Mflv_3847|M.gilvum_PYR-GCK SDEAMLDAILAGASGYVVKDIKGMELARAIKEVGSGRSLLDNRAAAALMA
TH_2159|M.thermoresistible__bu SDQAMLDAILAGASGYVVKDIKGMELAQAIKDVGAGRSLLDNRAAEALMA
MMAR_3480|M.marinum_M SDEAMLEAILAGASGYVVKDIKGMELARAIKDVGAGKSLLDNRAAAALMA
MAV_4109|M.avium_104 SDEAMLEAILAGASGFVIKDIKGMELARAIKDVGAGKSLLDNRAAAALMA
MAB_3891c|M.abscessus_ATCC_199 DEHAMLDAIMAGAGGYVIKDIKGMELISAVRTVGAGRSLLDNRAAAALMN
MLBr_01286|M.leprae_Br4923 QRDLVERARDAGAMAYLVKPFTISDLIPAIALAMSRFSELT-----ALER
. . : * *** .:::* . :* *: : * : *:
MSMEG_6236|M.smegmatis_MC2_155 SLSG---RTEPREQLTDREIEVLRLLAAGLSNNKIGEKLFISATTAKFHV
Mvan_5474|M.vanbaalenii_PYR-1 SMSG---RSESRAQLTDREVEVLRLLAAGLSNNKIGERLYISATTAKFHV
Mb3157c|M.bovis_AF2122/97 KLRGAAEKQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYV
Rv3133c|M.tuberculosis_H37Rv KLRGAAEKQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYV
MUL_2423|M.ulcerans_Agy99 RLRGVVERPDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYV
Mflv_3847|M.gilvum_PYR-GCK KLRGEGERTDPLSGLTDQERVLLDLLGEGLTNKQIAARMFLAEKTVKNYV
TH_2159|M.thermoresistible__bu KLRGADHQPDPLAGLSDQERVLLSLIGEGLTNKQIAARMFLAEKTVKNYV
MMAR_3480|M.marinum_M KLRGGAEQKDPLSGLTEQERTLLGHLSEGLTNRQIAARMFLAEKTVKNYV
MAV_4109|M.avium_104 KLRGSAAQADPLSGLTEQERTLLGLLSEGLTNRQIAARMFLAEKTVKNYV
MAB_3891c|M.abscessus_ATCC_199 RLRAAADNPSPLAGLTAQERTLLELIGEGLTNRQIAERMFLAEKTVKNYV
MLBr_01286|M.leprae_Br4923 EVATLADRLETR-KLVERAKGLLQVK-QGMTEPEAFKWIQRAAMDRRTTM
: . .. * : :* *::: : : : : :
MSMEG_6236|M.smegmatis_MC2_155 SNIMRKLDVSRRAEAVYAASKRGLI------------
Mvan_5474|M.vanbaalenii_PYR-1 SNIMRKLEVSRRAEAVYAASKRGLI------------
Mb3157c|M.bovis_AF2122/97 SRLLAKLGMERRTQAAVFATELKR--SRPPGDGP---
Rv3133c|M.tuberculosis_H37Rv SRLLAKLGMERRTQAAVFATELKR--SRPPGDGP---
MUL_2423|M.ulcerans_Agy99 SRLLAKLGMERRTQAAVFAAELKR--ARSAGHR----
Mflv_3847|M.gilvum_PYR-GCK SRLLAKLGMERRTQAAVFISKLDR--GHGAGNA----
TH_2159|M.thermoresistible__bu SRLLAKLGMERRTQAAVFVSKLDR--PIDPRG-----
MMAR_3480|M.marinum_M SRLLAKLGMERRTQAAVFASRLGQ--GTRPVQGD---
MAV_4109|M.avium_104 SRLLAKLGMERRTQAAVFASKLNQQAGRPPTPPDWPG
MAB_3891c|M.abscessus_ATCC_199 SRLLTKLDLERRTQVAVLATKLAR--ENQGLHG----
MLBr_01286|M.leprae_Br4923 KRVAEVVLETLDTPKDT--------------------
..: : :