For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNLLAQQDFSTPWSEVHTLLLPAYGETWIMVGVTMTLVVLAGIPLGVVLHNTSEFGLRPNPSVFRVLNTM VNIGRSLPFLILMAAIIPVTRFLVGTTVGIPAAIVPMTLAGVPFFARLVQNALREVNSDVADMGLVSGGN LLQVVRTVQLSEALPALAGALTVNVIAMIEYSAIAGSIGAGGVGNLAITYGYNRFDDNIMIATAVSLIIT IQVVQFAGDRLVKALTR
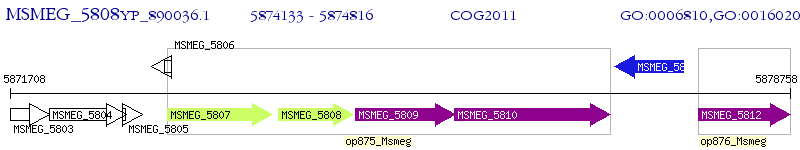
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5808 | - | - | 100% (227) | binding-protein-dependent transport systems inner membrane component |
| M. smegmatis MC2 155 | MSMEG_3057 | - | 1e-40 | 43.15% (197) | ABC-type metal ion transport system, permease component |
| M. smegmatis MC2 155 | MSMEG_0646 | - | 4e-10 | 24.49% (196) | putative transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 1e-06 | 23.23% (198) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_1209 | - | 1e-09 | 27.69% (195) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3757c | proW | 1e-06 | 23.23% (198) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2802c | - | 3e-35 | 36.53% (219) | ABC transporter permease |
| M. marinum M | MMAR_5299 | proZ | 3e-06 | 26.11% (157) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. avium 104 | MAV_0319 | - | 2e-06 | 27.15% (151) | ABC transporter, permease protein |
| M. thermoresistible (build 8) | TH_2403 | - | 4e-67 | 56.22% (217) | binding-protein-dependent transport systems inner membrane |
| M. ulcerans Agy99 | MUL_4375 | proW | 2e-06 | 23.27% (202) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_3885 | - | 1e-103 | 81.94% (227) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3783c|M.bovis_AF2122/97 -----------------MHYLMTHPGAAWALTVVHLRLS-LLPVLIGLMS
Rv3757c|M.tuberculosis_H37Rv -----------------MHYLMTHPGAAWALTVVHLRLS-LLPVLIGLMS
MUL_4375|M.ulcerans_Agy99 -----------------MHYLMTHLGTAWALTMVHLRLS-LIPVLIGTAI
MAV_0319|M.avium_104 -----------------MHYLLTHLDDAARLTVVHLRLS-LVPVLIGLAI
Mflv_1209|M.gilvum_PYR-GCK -----------------MRYLFTHLDDLWVLTLIHLRLS-LIPIVLGLLI
MSMEG_5808|M.smegmatis_MC2_155 -MNLLAQQDFSTPWSEVHTLLLPAYGETWIMVGVTMTLVVLAGIPLGVVL
Mvan_3885|M.vanbaalenii_PYR-1 -MNYLALDDFSTPWLKVPDLLLPAYGETWVMVGVTMVLVVLIGTPAGIIL
TH_2403|M.thermoresistible__bu VTSVLAAG--TVPLDEMAGLLFPALLDTLTMVGIVMTIVTLVGVPLGALV
MAB_2802c|M.abscessus_ATCC_199 ---------MTTNWERLRPQLFEAFGDTLYMVSITLVVGGLVGLALGVLL
MMAR_5299|M.marinum_M ----------MNFLQQALSYLSTAANWTGSAGLAHRTAEHLEYTALAVTA
* * .
Mb3783c|M.bovis_AF2122/97 AVPLGLLVQRAPLLRRLTTATASVIFTIPSLALFVVLP---LIIGTRILD
Rv3757c|M.tuberculosis_H37Rv AVPLGLLVQRAPLLRRLTTATASVIFTIPSLALFVVLP---LIIGTRILD
MUL_4375|M.ulcerans_Agy99 AIPLGLFVQHAPTLRRLTTATASVVFTIPSLALFVVLP---LIIKMRILQ
MAV_0319|M.avium_104 ALPLGMLVQRRRFARRLTTAVASVVFTIPSLALFVVLP---MIIGTRILD
Mflv_1209|M.gilvum_PYR-GCK AVPLGALVQRTTALRRLTTVTASIIFTIPSLALFVVLP---LIIPTRILD
MSMEG_5808|M.smegmatis_MC2_155 HNTSEFGLRPNPSVFRVLNTMVNIGRSLPFLILMAAIIPVTRFLVGTTVG
Mvan_3885|M.vanbaalenii_PYR-1 HNTSTLGLFPNPRVFTVLNTVVNVGRSLPFLILMAAIIPVTRFLVGTTIG
TH_2403|M.thermoresistible__bu HNLAPGGLFENPTLHTVLSWTINIGRSLPFLILMAAIMPFTRFVTGTNIG
MAB_2802c|M.abscessus_ATCC_199 YTTRSGGLLQNRLLHTLLNVVVNIVRPIPFVVLIAALGPITLGVVGTTIG
MMAR_5299|M.marinum_M SALVAIPIG--LIIGHTGRGQLLVVSAVNGLRALPTLGVLLLGVLLFGLG
: : .: : : .: : :
Mb3783c|M.bovis_AF2122/97 EANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQMLKVE
Rv3757c|M.tuberculosis_H37Rv EANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQMLKVE
MUL_4375|M.ulcerans_Agy99 EANVIIALSAYTTALLVRAVLEALDAVPAQVRDAAVAVGYSPIARMLKVE
MAV_0319|M.avium_104 EANVMVALTAYTAALLVRAVLEALDAVPAQISDAATAVGYPRLTRMMKVE
Mflv_1209|M.gilvum_PYR-GCK EANVIVALTLYTVALLVRAVPEALDAVSPAVLDAATAVGYRPLTRMLKVE
MSMEG_5808|M.smegmatis_MC2_155 IPAAIVPMTLAGVPFFARLVQNALREVNSDVADMGLVSGGNLLQVVRTVQ
Mvan_3885|M.vanbaalenii_PYR-1 IAAAIVPMTVAGIPFFARLVQNALREVRSDVTDMGLVSGGSHVQVIRSVQ
TH_2403|M.thermoresistible__bu IAAAVVPMSLAGIAFFTRIVENSLRSVPPSVVQVARASGGSPLQIIRTAQ
MAB_2802c|M.abscessus_ATCC_199 TTAVVFVMIVAASFAIARIVEQNLVTIDPGVIEAARAVGAGPLRIIVTLL
MMAR_5299|M.marinum_M LGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTEAQVVLRVE
:. : : . : : . : : . . * :
Mb3783c|M.bovis_AF2122/97 LPLSIPVLVAGLRVVAVTNIAMVSVGSVIGIGGLG-TWFTAGYQTNKSDQ
Rv3757c|M.tuberculosis_H37Rv LPLSIPVLVAGLRVVAVTNIAMVSVGSVIGIGGLG-TWFTAGYQTNKSDQ
MUL_4375|M.ulcerans_Agy99 LPLSIPVLVSGLRVVVVTNIAMVSVGAVIGIGGLG-TWFTAGYLMDKSDQ
MAV_0319|M.avium_104 LPLAVPVLIAGLRVVVVTNIAMVSVGSVIGIGGLG-SWFTQGYQTGKSDQ
Mflv_1209|M.gilvum_PYR-GCK LPLALPVLIASLRVVAVTNISMVAVGSVIGIGGLG-TWFTEGYQANKSDQ
MSMEG_5808|M.smegmatis_MC2_155 LSEALPALAGALTVNVIAMIEYSAIAGSIGAGGVGNLAITYGYNRFDDNI
Mvan_3885|M.vanbaalenii_PYR-1 LSEALPALAGALTVNTIAMIEYSAIAGSIGAGGVGHLAITYGYNRFDDNI
TH_2403|M.thermoresistible__bu LHEAVPSILGGLTINVIAMIEYSAIAGTIGAGGIGYVAVTYGYQRFDQGV
MAB_2802c|M.abscessus_ATCC_199 VPEALGPLILGYTFVLIGIVDMSAMAGLVGGGGLGDFAIVQGYQRFNWQV
MMAR_5299|M.marinum_M MPNALPLILGGLRSATLQVVATATVAAYASLGGLG-RYLIDGIKERQFHI
: :: : . : : ::.. . **:* . * .
Mb3783c|M.bovis_AF2122/97 IVAGVVAMFLLAIVVDVVINLAGRLATP--------WERAPRAARRRRQV
Rv3757c|M.tuberculosis_H37Rv IVAGVVAMFLLAIVVDVVINLAGRLATP--------WERAPRAARRRRQV
MUL_4375|M.ulcerans_Agy99 IVAGITVMFVLAILIDMVLNVAGRLATP--------WERSSRGRRRFR--
MAV_0319|M.avium_104 ILAGIIALFALAVAVDGLIVLGGRLVTP--------WAHAVRGGARRS-V
Mflv_1209|M.gilvum_PYR-GCK IIAGIVAIFVLAIVIDTLIMWLGKLLTP--------WTRAQPRSTRTQ--
MSMEG_5808|M.smegmatis_MC2_155 MIATAVSLIITIQVVQFAGDRLVKALTR----------------------
Mvan_3885|M.vanbaalenii_PYR-1 MIATVIALIITIQLVQFTGDRVVRALTR----------------------
TH_2403|M.thermoresistible__bu MAATIVILVCTVAVVQLIGDALVRFTTP--------HARARAARV-----
MAB_2802c|M.abscessus_ATCC_199 TIVSTVIIIALVQAAQFFGNYLARKALR--------R-------------
MMAR_5299|M.marinum_M ALVGALMVAALALSLDGLLALAGWASVPGTGRLSRFWHQSDDRIDVAVSP
. : :
Mb3783c|M.bovis_AF2122/97 AAPITGGAR--
Rv3757c|M.tuberculosis_H37Rv AAPITGGAR--
MUL_4375|M.ulcerans_Agy99 -SPIVGGAR--
MAV_0319|M.avium_104 VAPVVGGAR--
Mflv_1209|M.gilvum_PYR-GCK ---KAGASA--
MSMEG_5808|M.smegmatis_MC2_155 -----------
Mvan_3885|M.vanbaalenii_PYR-1 -----------
TH_2403|M.thermoresistible__bu -----------
MAB_2802c|M.abscessus_ATCC_199 -----------
MMAR_5299|M.marinum_M NSAAVGSHVLR