For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NFLQQALSYLSTAANWTGSAGLAHRTAEHLEYTALAVTASALVAIPIGLIIGHTGRGQLLVVSAVNGLRA LPTLGVLLLGVLLFGLGLGPPIVALMLLGMPPLLAGTYAGIASVDPLVVDAARAMGMTEAQVVLRVEMPN ALPLILGGLRSATLQVVATATVAAYASLGGLGRYLIDGIKERQFHIALVGALMVAALALSLDGLLALAGW ASVPGTGRLSRFWHQSDDRIDVAVSPNSAAVGSHVLR*
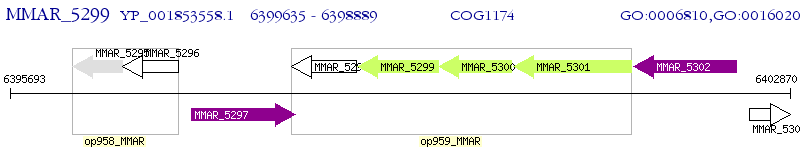
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5299 | proZ | - | 100% (248) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) transport integral membrane protein ABC transporter ProZ |
| M. marinum M | MMAR_0033 | - | 5e-31 | 37.57% (189) | hypothetical protein MMAR_0033 |
| M. marinum M | MMAR_0035 | - | 2e-17 | 30.93% (194) | ABC transporter permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 1e-107 | 80.65% (248) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_1210 | - | 6e-94 | 72.18% (248) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 1e-107 | 80.65% (248) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0264 | - | 4e-90 | 75.91% (220) | osmoprotectant ABC transporter ProZ |
| M. avium 104 | MAV_0320 | - | 5e-99 | 78.79% (231) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_6331 | - | 2e-91 | 75.45% (220) | ABC transporter, permease protein ProZ |
| M. thermoresistible (build 8) | TH_3904 | - | 5e-32 | 36.51% (189) | PUTATIVE conserved hypothetical membrane protein |
| M. ulcerans Agy99 | MUL_4374 | proZ | 1e-131 | 97.58% (248) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_5598 | - | 1e-89 | 74.32% (222) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5299|M.marinum_M ------------------------------------------MNFLQQAL
MUL_4374|M.ulcerans_Agy99 ------------------------------------------MNFLQQAL
Mb3782c|M.bovis_AF2122/97 ------------------------------------------MNFLQQAL
Rv3756c|M.tuberculosis_H37Rv ------------------------------------------MNFLQQAL
MAV_0320|M.avium_104 ------------------------------------------MNFVQRAI
Mflv_1210|M.gilvum_PYR-GCK ------------------------------------------MNFLQEAL
Mvan_5598|M.vanbaalenii_PYR-1 ------------------------------------------MNFLQEAL
MSMEG_6331|M.smegmatis_MC2_155 ------------------------------------------MNFLQQAL
MAB_0264|M.abscessus_ATCC_1997 ------------------------------------------MNFLSNAL
TH_3904|M.thermoresistible__bu VTATVDDTGDTGHTGCTGTAGDRASRSAARRWARRTVQPVLCLVAVGGAL
: : *:
MMAR_5299|M.marinum_M SYLSTAAN------WTGSAGLAHRTAEHLEYTALAVTASALVAIPIGLII
MUL_4374|M.ulcerans_Agy99 SYLSTAAN------WTGSAGLAHRTAEHLEYTALAATASALVAIPIGLII
Mb3782c|M.bovis_AF2122/97 SYLLTASN------WTGPVGLAVRTCEHLEYTAVAVAASALIAVPVGLLI
Rv3756c|M.tuberculosis_H37Rv SYLLTASN------WTGPVGLAVRTCEHLEYTAVAVAASALIAVPVGLLI
MAV_0320|M.avium_104 AYLLSADN------WTGPVGLAARILEHLEYTGIAVGASALIAVPVGLLI
Mflv_1210|M.gilvum_PYR-GCK SFIFTAAN------WAGPAGLTARILEHLQYTVIAVFFSALIAVPLGMLI
Mvan_5598|M.vanbaalenii_PYR-1 SFIFTAAN------WGGPAGLSARILEHLQYTVIAVFFSAVIAVPLGMLI
MSMEG_6331|M.smegmatis_MC2_155 AFIFTAEN------WAGPSGLGARIVEHLEYTAVAVFFSALIAVPLGMVI
MAB_0264|M.abscessus_ATCC_1997 GYIFTASH------WAGSSGLGVRIGEHLEYTALAVLAAVVIAVPLGLFI
TH_3904|M.thermoresistible__bu AYVHLADVSESEKRSLGVPHLLMLLRQHMEISLAATVLTCAIAIPLGVAL
.:: * * * :*:: : *. : :*:*:*: :
MMAR_5299|M.marinum_M G--HTGRGQLLVVSAVNGLRALPTLGVLLLGVLLFGLGLGPPIVALMLLG
MUL_4374|M.ulcerans_Agy99 G--HTGRGQLLVVSAVNGLRALPTLGVLLLGVLLFGLGLGPPIVALMLLG
Mb3782c|M.bovis_AF2122/97 G--HTGRGTLLVVGAVNGLRALPTLGVLLLGVLLFGLGLGPPLVALMLLG
Rv3756c|M.tuberculosis_H37Rv G--HTGRGTLLVVGAVNGLRALPTLGVLLLGVLLFGLGLGPPLVALMLLG
MAV_0320|M.avium_104 G--HTGRGTLLVVGAVNGLRALPTLGVLLFGTLLFGLGLGPSLVALMLLG
Mflv_1210|M.gilvum_PYR-GCK G--HTGRGTFVVVTGVNALRALPTLGVLLLGVLLWGLGLIPPTVALMLLG
Mvan_5598|M.vanbaalenii_PYR-1 G--HTGRGTFVVVTGVNALRALPTLGVLLLGVLLWGLGLIPPTVALMLLG
MSMEG_6331|M.smegmatis_MC2_155 G--HTGRGTFLVVTGVNALRALPTLGVLLLGVLLWGLGLVPPTVALMLLG
MAB_0264|M.abscessus_ATCC_1997 G--HTGRGIFLVVSAVNALRALPTLGMLLLGVLLWGLGLVPPTVALLLLG
TH_3904|M.thermoresistible__bu TRGRFRRFATPIITVSGFGQAAPAIGLIALGAVLFGIGAVGAVAALTVYG
: * :: . :* *::*:: :*.:*:*:* . .** : *
MMAR_5299|M.marinum_M MPPLLAGTYAGIASVDPLVVDAARAMGMTEAQVVLRVEMPNALPLILGGL
MUL_4374|M.ulcerans_Agy99 MPPLLAGTYAGIASVDPLVVDAARAMGMTEAQVVLRVEMPNALPLILGGL
Mb3782c|M.bovis_AF2122/97 IPSLLASTYAGIASVDPLVVDAARAMGMTESQVLLRVEVPNALPLMLGGL
Rv3756c|M.tuberculosis_H37Rv IPSLLASTYAGIASVDPLVVDAARAMGMTESQVLLRVEVPNALPLMLGGL
MAV_0320|M.avium_104 VPALLAGTYAGIANVEPTVVDAARAMGMTESQVLLRVEVPNALPLILGGL
Mflv_1210|M.gilvum_PYR-GCK IPPLLAGTYSGIANVDRSVVDAARSMGMTESRILLRVETPNALPLILGGL
Mvan_5598|M.vanbaalenii_PYR-1 IPPLLAGTYSGIANVDRDVVDAARAMGMTESRILLRVETPIAMPLILGGL
MSMEG_6331|M.smegmatis_MC2_155 IPPLLAGTYAGVANVERTVVDAARSMGMTESRILLRVEVPNALPLILGGL
MAB_0264|M.abscessus_ATCC_1997 VPPLLAGTYSGIANVDRTVVEAARAMGMTERQVLFGVEVPNALPLILSGL
TH_3904|M.thermoresistible__bu ALPIIANTVTGLNGVDDRLIEAARGMGMPTSTTLLRVELPLALPVIIAGV
.::*.* :*: .*: :::***.***. :: ** * *:*:::.*:
MMAR_5299|M.marinum_M RSATLQVVATATVAAYASLGGLGRYLIDGIKERQFHIALVGALMVAALAL
MUL_4374|M.ulcerans_Agy99 RSATLQVVATVTVAAYASLGGLGRYLIDGIKERQSHIALVGALMVAALAL
Mb3782c|M.bovis_AF2122/97 RSATLQVVATATVAAYASLGGLGGYLIDGIKERRFHIALVGAMMVAALAL
Rv3756c|M.tuberculosis_H37Rv RSATLQVVATATVAAYASLGGLGGYLIDGIKERRFHIALVGAMMVAALAL
MAV_0320|M.avium_104 RSATLQVVATATVAAYASLGGLGRYLIDGIKEREFHLALVGALTVAALAL
Mflv_1210|M.gilvum_PYR-GCK RTATLQIVATATVAAYASLGGLGRYLIDGIKVREFYLALVGALMVTALAL
Mvan_5598|M.vanbaalenii_PYR-1 RTATLQIVATATVAAYASLGGLGRYLIDGIKVREFYLALVGALMVTALAL
MSMEG_6331|M.smegmatis_MC2_155 RTSTLQIVATATVAAYASLGGLGRYLIDGIKIREFHIALVGAIMVTALAL
MAB_0264|M.abscessus_ATCC_1997 RTATLQVVATATVAAYASLGGLGRYLIDGIKVRQFHLALVGAIMVTALAL
TH_3904|M.thermoresistible__bu RTALVLIVGTAALASFTGAGGLGQLITTGIKLQQTVTLIVGAVLVAALAL
*:: : :*.*.::*:::. **** : *** :. :***: *:****
MMAR_5299|M.marinum_M SLDGLLALAGWASVPGTGRLSRFWH------QSDDRIDVAVSPNSAAVGS
MUL_4374|M.ulcerans_Agy99 SLDGLLALAGWASVPGTGRVSRFWH------QPDDRIDVAVSANSAAVGS
Mb3782c|M.bovis_AF2122/97 TLDGLLALAGWVSVPGTGRMRKL---------------AAVVDKPAAGGG
Rv3756c|M.tuberculosis_H37Rv TLDGLLALAGWVSVPGTGRMRKL---------------AAVVDKPAAGGG
MAV_0320|M.avium_104 VLDGLLAVAVWASVPGTGRLRRT---------------YRPLPHRLAGTS
Mflv_1210|M.gilvum_PYR-GCK ILDALLAFAVWASVPGTGRLSRRRA----MPQPFLDDEVALESGSPAVRS
Mvan_5598|M.vanbaalenii_PYR-1 ILDAALAFAVWASVPGSGRLHRSGS----MPQPLLGDEVALESRSRAGRR
MSMEG_6331|M.smegmatis_MC2_155 ILDAALAFAVWLSVPGAGRLRGRRSGRGQMPQPFLGDEVALESRTPNASA
MAB_0264|M.abscessus_ATCC_1997 LLDAVLALAVWLSAPGTGRLRPVPD------LPDREGHAVYADSGVSAGA
TH_3904|M.thermoresistible__bu FIDWLARMVEVVAAPKGL--------------------------------
:* .. :.*
MMAR_5299|M.marinum_M HVLR--------------
MUL_4374|M.ulcerans_Agy99 HVLR--------------
Mb3782c|M.bovis_AF2122/97 HALR--------------
Rv3756c|M.tuberculosis_H37Rv HALR--------------
MAV_0320|M.avium_104 YGR---------------
Mflv_1210|M.gilvum_PYR-GCK PVTNGGSRRIR-------
Mvan_5598|M.vanbaalenii_PYR-1 PDSEARHERGNPSPTVEA
MSMEG_6331|M.smegmatis_MC2_155 RSSGVGYEGGAPSHTVEG
MAB_0264|M.abscessus_ATCC_1997 MPRSPG------------
TH_3904|M.thermoresistible__bu ------------------