For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRHREFDVDKALDAALCVFWRKGYEGASYSDLTQATGVERPALYSAFGNKEALFLRALDRYYERYMDFI PVALEQPTSREVAEHLLRSAADLNTRYPDHTGCLGVQGCLVGTDDSEPIRRALVDARASGEASLRARFED AHATGDLPKSANPAALAAFLMTVLHGMAVQAKAGFSRETLDAIIDQALSSWPTPDRGSAGKATRKR
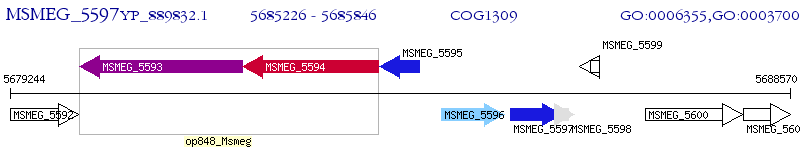
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5597 | - | - | 100% (206) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2030 | - | 1e-28 | 38.02% (192) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1509 | - | 3e-20 | 35.36% (181) | TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0149 | - | 3e-07 | 25.32% (158) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0944 | - | 1e-20 | 32.81% (192) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0144 | - | 3e-07 | 25.32% (158) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00815 | - | 2e-05 | 26.45% (121) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1149 | - | 4e-07 | 35.29% (68) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0630 | - | 4e-12 | 29.41% (204) | transcriptional regulatory protein |
| M. avium 104 | MAV_1611 | - | 2e-10 | 29.34% (167) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0251 | - | 5e-07 | 27.75% (173) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_4635 | - | 2e-06 | 32.39% (71) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1617 | - | 4e-46 | 48.44% (192) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0149|M.bovis_AF2122/97 MPHSWTPTSVMTPPLVVAAFRPVGHYRLATDRAGGPCSPPATGAKLTSSV
Rv0144|M.tuberculosis_H37Rv MPHSWTPTSVMTPPLVVAAFRPVGHYRLATDRAGGPCSPPATGAKLTSSV
MSMEG_5597|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1617|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0944|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0630|M.marinum_M --------------------------------------------------
MAB_1149|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_4635|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00815|M.leprae_Br4923 -------------------------------------------------M
MAV_1611|M.avium_104 --------------------------------------------------
TH_0251|M.thermoresistible__bu --------------------------------------------------
Mb0149|M.bovis_AF2122/97 ASRPTVGTKPQWWHTLVMSMSLTAGRGPGRPPAAKADETRKRILHAARQV
Rv0144|M.tuberculosis_H37Rv ASRPTVGTKPQWWHTLVMSMSLTAGRGPGRPPAAKADETRKRILHAARQV
MSMEG_5597|M.smegmatis_MC2_155 ---------------------------MGRHREFDVDK----ALDAALCV
Mvan_1617|M.vanbaalenii_PYR-1 -------------------MGSPGTSSIGRPRSFDTDE----ALERAMVV
Mflv_0944|M.gilvum_PYR-GCK ---------------------------MARPRTFDEDE----VIEAGRDL
MMAR_0630|M.marinum_M ---------------------------MARPRAFDEAV----VLSAARDE
MAB_1149|M.abscessus_ATCC_1997 ----------------MAVPSRPDVPGRTPRVRMAGSERRRQLIDVARSL
MUL_4635|M.ulcerans_Agy99 ---------------------------------MTGSERRHQLIGIARSL
MLBr_00815|M.leprae_Br4923 NNLAKAAQRRTVRPIDRGRLSDDIATPNQRSNRLSRDKRRGQLLIVASDV
MAV_1611|M.avium_104 -------------------MLLGMSPVKPLRSRPTRGEVRDRILDAAAKV
TH_0251|M.thermoresistible__bu --------------------------VTDDHLAARRRQ----ILDGARRC
: .
Mb0149|M.bovis_AF2122/97 FSERGYDGATFQEIAVRADLTRPAINHYFANKRVLYQEVVEQTHELVIVA
Rv0144|M.tuberculosis_H37Rv FSERGYDGATFQEIAVRADLTRPAINHYFANKRVLYQEVVEQTHELVIVA
MSMEG_5597|M.smegmatis_MC2_155 FWRKGYEGASYSDLTQATGVERPALYSAFGNKEALFLRALDRYYERYMDF
Mvan_1617|M.vanbaalenii_PYR-1 FWAHGYEGASLTDLTEAMGITRTSMYAAFGNKEELFRAALTRYLEGPGGY
Mflv_0944|M.gilvum_PYR-GCK FWSQGYAATSIDDLSAATGLGRGSLYKAFGDKHSMFLRGLEQYCTEAIAG
MMAR_0630|M.marinum_M FRSHGYAATSVDSLAAATGLNRSSLYGSFGDKHRLFLRALDGYCEATLHD
MAB_1149|M.abscessus_ATCC_1997 FAERGYEATSIEEIAQRAGVSKPVVYEHFGGKEGLYAVVVDREMSVLLDG
MUL_4635|M.ulcerans_Agy99 FAERGYDGTSIEEIAQRANVSKPVVYEHFGGKEGLYAVVVDREMSALLDG
MLBr_00815|M.leprae_Br4923 FVDHGYHAAGMDEIADRAGVSKPVLYQHFSSKLELYLAVLERHMNNLVSS
MAV_1611|M.avium_104 FAAEGFAGATIDAIGQAAGFTKGAVYSNFESKDELFLALLDREFELRGQQ
TH_0251|M.thermoresistible__bu FAEYGYEQATVRRLEQATGLSRGAIFHHFKDKDTLFFELAREDAERMAEV
* *: : : .. : : * .* ::
Mb0149|M.bovis_AF2122/97 GIERARREP--TLMGRLAVVVDFAMEADAQYPASTAFLAT--TVLESQRH
Rv0144|M.tuberculosis_H37Rv GIERARREP--TLMGRLAVVVDFAMEADAQYPASTAFLAT--TVLESQRH
MSMEG_5597|M.smegmatis_MC2_155 -IPVALEQP--TSREVAEHLLRSAADLNTRYPDHTGCLGVQGCLVGTDDS
Mvan_1617|M.vanbaalenii_PYR-1 -LSRALEES--TAREVAEAFLRGSVRATTRPEGPAGCLVVQGSLAASDAG
Mflv_0944|M.gilvum_PYR-GCK -IRAELRDQDLSAYDRLVAHVRGRAADVVSDNDLRGCLLAKSAAELSSTD
MMAR_0630|M.marinum_M -VREVLRERGVSARQRLINHVHAIVNGIVADTDRRGCMMSRSSAELAGAD
MAB_1149|M.abscessus_ATCC_1997 -ITSSLTSY--RSRVRLEQVALALLSYIEERTDGFRILIRDSPPGVSSG-
MUL_4635|M.ulcerans_Agy99 -ITSSLTNN--RSRVRVERVALALLTYVEERTDGFRIMIRDSPASISSG-
MLBr_00815|M.leprae_Br4923 -VQQALRTT-TNNRQRLHAAVQAFFDFIEHDGQGYRLIFENDNITEPRVA
MAV_1611|M.avium_104 -IAIALDRSAGDTTAAAREVSRSVLDSVRDHSDYYVLLVEYWLRAQRDP-
TH_0251|M.thermoresistible__bu -----------ASREGLIQVMRDLIAAPEQYDWLTTRLEIARKLRNDPE-
:
Mb0149|M.bovis_AF2122/97 PELSRTENDAVRATREFLVWAVNDAIERGELAADVDVSSLAETLLVVLCG
Rv0144|M.tuberculosis_H37Rv PELSRTENDAVRATREFLVWAVNDAIERGELAADVDVSSLAETLLVVLCG
MSMEG_5597|M.smegmatis_MC2_155 EPIRRALVDARASGEASLRARFEDAHATGDLPKSANPAALAAFLMTVLHG
Mvan_1617|M.vanbaalenii_PYR-1 QPVRDALIACREEAVSRLRERFRRAVKDGDLPPGTDPGLLARYLMTVGNG
Mflv_0944|M.gilvum_PYR-GCK PEVARHVKRTLDAWLRELTATISAAQADGDIPSSTEAKALAALILAVLRG
MMAR_0630|M.marinum_M PDVSGIVERSLEAWRRELADCIAEAQLEGAVAGDGSPQALATVMLSLMQG
MAB_1149|M.abscessus_ATCC_1997 -TYSSLLNDAVSQVSSILAGDFQRRGLDAEMAPLYAQALVGQVSMTAQWW
MUL_4635|M.ulcerans_Agy99 -TYSSLLNDAVSQVSSILAGDFARRGLDSELAPLYAQALVGSVSMTAQWW
MLBr_00815|M.leprae_Br4923 AQVRVATESCIDAVFALISADSR---LDPHRARMVAVGLVGISVDCARYW
MAV_1611|M.avium_104 ----QLRERLIERRRAAADQALHIVESTDTVPGDRRLTDIAQLVVTLNLG
TH_0251|M.thermoresistible__bu --FHRGWSELSAELAAATTARLRRQKQAGRLRDDVPGEVLRTYLELVLDG
:
Mb0149|M.bovis_AF2122/97 VGFYI--GFVGSYQR-MATITDSFQQLLAGTLWRPPT------
Rv0144|M.tuberculosis_H37Rv VGFYI--GFVGSYQR-MATITDSFQQLLAGTLWRPPT------
MSMEG_5597|M.smegmatis_MC2_155 MAVQA--KAGFSRET-LDAIIDQALSSWPTPDRGSAGKATRKR
Mvan_1617|M.vanbaalenii_PYR-1 IAVQA--ASGVARRE-LQRVADAALRHWPPA------------
Mflv_0944|M.gilvum_PYR-GCK MEALR--KGGASAAT-VKATAEQLVAML---------------
MMAR_0630|M.marinum_M FEALG--KGGVGSAE-LRAAESAALALLFGAGTDPKP------
MAB_1149|M.abscessus_ATCC_1997 LDAREP-KKEVVAAH-IVNLCWHGLKHLEADPQLHEE------
MUL_4635|M.ulcerans_Agy99 LDAREP-KKEVVAAH-LVNLVWNGLTHLEADPQLQDE------
MLBr_00815|M.leprae_Br4923 LDTNRP-ISKSDAVEGTVQFAWGGLSHVPLTRL----------
MAV_1611|M.avium_104 VAMEEVLRPGTINPDLLAQLITALLESIPV-------------
TH_0251|M.thermoresistible__bu LVARLASGDDPEKLSAVLDLVEASVRHQGSE------------
.