For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PMTGQFGRVDRRPLYEQVADRLREFIDTSGMQPGDKLANVRDLAAELQVGRSSIREAVNALRAQRIVEVR HGDGIYLLQRPEDVIESLATELVETHVDHPYIWETRQALETQCARLAAVHATDDDLRELDASLDQMRAEV EKGLPGLEGDRRFHLGVAVAAHNPLLLRLLKGLRVALDRTSETSLTRPGQPARSLEEHRGVVEAIRAGDA ADAANKMLAHLVGTTDELVRR*
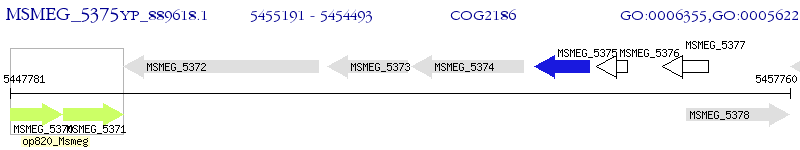
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5375 | - | - | 100% (232) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2173 | - | 7e-37 | 38.60% (228) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2794 | - | 5e-25 | 36.89% (206) | transcriptional regulator, GntR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 2e-11 | 25.45% (224) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0016 | - | 7e-17 | 32.58% (221) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0494 | - | 3e-11 | 25.45% (224) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4644c | - | 1e-13 | 28.51% (221) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0749 | - | 1e-14 | 30.36% (224) | GntR family transcriptional regulator |
| M. avium 104 | MAV_1576 | - | 1e-11 | 28.14% (231) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_4115 | - | 2e-11 | 28.43% (204) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1381 | - | 3e-14 | 31.25% (224) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0294 | - | 3e-17 | 34.10% (217) | GntR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0749|M.marinum_M ----------MQTVRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
MUL_1381|M.ulcerans_Agy99 -------------MRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
Mflv_0016|M.gilvum_PYR-GCK MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVE
MAB_4644c|M.abscessus_ATCC_199 ---------MHPRLSSQSLAEQVANILAAQIGDGRWSTGAKLPGEVALCE
Mb0505|M.bovis_AF2122/97 MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
Rv0494|M.tuberculosis_H37Rv MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
MSMEG_5375|M.smegmatis_MC2_155 ----MPMTGQFGRVDRRPLYEQVADRLREFIDTSGMQPGDKLANVRDLAA
Mvan_0294|M.vanbaalenii_PYR-1 MNAPGGEAIPPLIRRPQSVSVALAAHFERLIATGEISPGTRLPSERELAA
TH_4115|M.thermoresistible__bu ---------------VQSSYIQVADQLRDLILRGDLAVGQKLPSEAEMAP
MAV_1576|M.avium_104 -------MSVPDFAARPQLSDDVAHLIRGRIFDGSYAAGSYVR-LDQLAA
: : : * : :
MMAR_0749|M.marinum_M MTGTARNTVREAVQALVHVGILERRQ---GSGTYVIA-----DVEQGSAL
MUL_1381|M.ulcerans_Agy99 MTGTARNTVREAVQALVHVGILERRQ---GSGTYVIA-----DVEQGSAL
Mflv_0016|M.gilvum_PYR-GCK KFGVGRNTVREAVRALAHAGILEVRQ---GDGTYVRA-----TSEVSGAL
MAB_4644c|M.abscessus_ATCC_199 EFGVSRATVREAIRDLKTKGLVSTRQ---GAGVFVTS-----DRPLG-QW
Mb0505|M.bovis_AF2122/97 RLGVNRTSLRQGLARLQQMGLIEVRH---GSGSVVRDPEGLTHPAVVEAL
Rv0494|M.tuberculosis_H37Rv RLGVNRTSLRQGLARLQQMGLIEVRH---GSGSVVRDPEGLTHPAVVEAL
MSMEG_5375|M.smegmatis_MC2_155 ELQVGRSSIREAVNALRAQRIVEVRH---GDGIYLLQ----RPEDVIESL
Mvan_0294|M.vanbaalenii_PYR-1 SMSVSRSSLREAMHELESKRLVSRTR---GRGTIVLAP----TASVDELL
TH_4115|M.thermoresistible__bu LLGVSRSTIREALRILTTEHLIETRRGIQGGAFVAVPDSERLEGMFNTSF
MAV_1576|M.avium_104 ELGISVTPVREALFALRAEGLISQQP---RRGFVVLP-------------
.:*:.: * ::. .
MMAR_0749|M.marinum_M AEYFAAARERD--LWELREAVEVTAAVLSARRRDAADIEDLRALLARRNE
MUL_1381|M.ulcerans_Agy99 AEYFAAARERD--LWELREAVEVTAAVLSARRRDAADIEDLRVLLARRNE
Mflv_0016|M.gilvum_PYR-GCK RR-LCGEELRE--VLEVRRALEVEAARLAASRRTPADVAEMRGLLARRDE
MAB_4644c|M.abscessus_ATCC_199 DRVLAAAEIDA--VVEIRRLLEVQAAGLAAERRNRSDVAAIRRALVARAK
Mb0505|M.bovis_AF2122/97 VRKLGPDFLVE--LLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQQ
Rv0494|M.tuberculosis_H37Rv VRKLGPDFLVE--LLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQQ
MSMEG_5375|M.smegmatis_MC2_155 ATELVETHVDHPYIWETRQALETQCARLAAVHATDDDLRELDASLDQMRA
Mvan_0294|M.vanbaalenii_PYR-1 AIAGGGTEQEH--AAELRSVIEPSIAELAAHRATSANVLQLRDVLDKSHE
TH_4115|M.thermoresistible__bu SMLALTNQVDARSFLQAMRALEGPAAALAAAARGGDQVEELKRTAAMVDP
MAV_1576|M.avium_104 ---VTRRDVTD--VANVQAHVGGELAARAAVNITDDQLRELKQIQAQLED
: : . :* : :
MMAR_0749|M.marinum_M LWSREPANDKERTEMTSADTALHRAIVAASHNEIYLQFYDLLLPTIHRAV
MUL_1381|M.ulcerans_Agy99 LWSREPANDKERTEMTSADTALHRAIVAASHNETYLQFYDLLLPTIHRAV
Mflv_0016|M.gilvum_PYR-GCK ---RDSAGDVE--AFARIDTEFHLAVVRCARNNTLIELYRGLLEAVTASV
MAB_4644c|M.abscessus_ATCC_199 -----AESAGDDIEYVDADLEFHRAVVAAVHNPLLDELFDSFSVRMREAM
Mb0505|M.bovis_AF2122/97 --------ADTAAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGRE
Rv0494|M.tuberculosis_H37Rv --------ADTAAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGRE
MSMEG_5375|M.smegmatis_MC2_155 -------EVEKGLPGLEGDRRFHLGVAVAAHNPLLLRLLKGLRVALDRTS
Mvan_0294|M.vanbaalenii_PYR-1 N--------LLQAESLRLDIEFHLLLAQAAQNPLLTALQTLASEWTGEVR
TH_4115|M.thermoresistible__bu ---------ADQEAAMQQSADFHSAILRASGNKLLEAMGRPVSAVARSKF
MAV_1576|M.avium_104 -----AYAGDEHERTVRLNHEFHRAINIAADSPKLAQLMSQITRYAPESV
. . : . . :
MMAR_0749|M.marinum_M EAG---PVGTCTSYEREHTELVEAIIAGDPQRAEAAAQVFIARLRP----
MUL_1381|M.ulcerans_Agy99 EAG---PVGTCTSYEREHTELVEAIIAGDPQRAEAAAQVFIARLRR----
Mflv_0016|M.gilvum_PYR-GCK AHTRHVPVDVC-----DHGVLLDHIAAGDPDLAARTAGEFLDRIINGLGS
MAB_4644c|M.abscessus_ATCC_199 LALLAGSAERP-HDQEHHQWLFDAIVDGSTRRAVELATDQLQHVRMR---
Mb0505|M.bovis_AF2122/97 HALTG-AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASALRMVK
Rv0494|M.tuberculosis_H37Rv HALTG-AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASALRMVK
MSMEG_5375|M.smegmatis_MC2_155 ETSLT-RPGQPARSLEEHRGVVEAIRAGDAADAANKMLAHLVGTTDELVR
Mvan_0294|M.vanbaalenii_PYR-1 KHSHSTRHGRCAS-VRGHQEIFDAVRTHDGLAARQAMEDHLRDVSELVAK
TH_4115|M.thermoresistible__bu RRTVP-GLEFWERNQRAHQELLKAILAGDENLAYELATRHIDEDLSPYYL
MAV_1576|M.avium_104 FPAIQ---GWPEQSMKDHRRILSALKKRDDKLARAAMSEHLAAGAVPLID
: : . * :
MMAR_0749|M.marinum_M -----------------
MUL_1381|M.ulcerans_Agy99 -----------------
Mflv_0016|M.gilvum_PYR-GCK ADSS-------------
MAB_4644c|M.abscessus_ATCC_199 -----------------
Mb0505|M.bovis_AF2122/97 SYRDRA-----------
Rv0494|M.tuberculosis_H37Rv SYRDRA-----------
MSMEG_5375|M.smegmatis_MC2_155 R----------------
Mvan_0294|M.vanbaalenii_PYR-1 ATRS-------------
TH_4115|M.thermoresistible__bu PPEEADRDGAGA-----
MAV_1576|M.avium_104 HLVARGVVVDGGGPDPG