For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNAPGGEAIPPLIRRPQSVSVALAAHFERLIATGEISPGTRLPSERELAASMSVSRSSLREAMHELESKR LVSRTRGRGTIVLAPTASVDELLAIAGGGTEQEHAAELRSVIEPSIAELAAHRATSANVLQLRDVLDKSH ENLLQAESLRLDIEFHLLLAQAAQNPLLTALQTLASEWTGEVRKHSHSTRHGRCASVRGHQEIFDAVRTH DGLAARQAMEDHLRDVSELVAKATRS
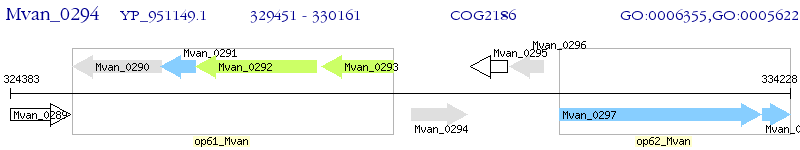
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0294 | - | - | 100% (236) | GntR domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_0606 | - | 1e-13 | 27.02% (248) | GntR domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_4590 | - | 2e-13 | 28.02% (257) | GntR domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 4e-10 | 26.34% (224) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2114 | - | 9e-14 | 28.02% (257) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0494 | - | 4e-10 | 26.34% (224) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4644c | - | 4e-15 | 30.70% (215) | GntR family transcriptional regulator |
| M. marinum M | MMAR_4029 | - | 3e-12 | 28.90% (218) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2560 | - | 2e-11 | 24.45% (229) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_2173 | - | 2e-22 | 33.50% (200) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_2873 | - | 4e-11 | 26.07% (234) | PUTATIVE regulatory protein GntR, HTH |
| M. ulcerans Agy99 | MUL_3894 | - | 3e-12 | 28.90% (218) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2560|M.avium_104 ----------------MEVTSLREP---KMADRVATVLRRMFIRGEITEG
TH_2873|M.thermoresistible__bu ------------VEDDMEVTSLREP---KMADRVATVLRRMFIRGEIPEG
Mflv_2114|M.gilvum_PYR-GCK MARSTPLAPMIGPDAVASRTAVRSP---KTAELVAGTLRRMVVEGQLKDG
Mb0505|M.bovis_AF2122/97 -----------MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPG
Rv0494|M.tuberculosis_H37Rv -----------MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPG
MMAR_4029|M.marinum_M ---------------MTDPEFAARP---QLSDDVARYVRKRIYEGNYAAG
MUL_3894|M.ulcerans_Agy99 ---------------MTDPEFAARP---QLSDDVARYVRKRIYEGNYAAG
MSMEG_2173|M.smegmatis_MC2_155 --------------MAEQLRPVTRP---RLYEVIVEQLCAYIYSNEMEPG
Mvan_0294|M.vanbaalenii_PYR-1 ---------MNAPGGEAIPPLIRRP--QSVSVALAAHFERLIATGEISPG
MAB_4644c|M.abscessus_ATCC_199 ----------------MHPRLSSQS----LAEQVANILAAQIGDGRWSTG
. :. . . . *
MAV_2560|M.avium_104 TMLPPESELMERFGVSRPTLREAFRVLESESLIQVQRGVRGGARVTRP-R
TH_2873|M.thermoresistible__bu TMLPPESELMERFGVSRPTLREAFRVLESESLIVVQRGVRGGARVTRP-R
Mflv_2114|M.gilvum_PYR-GCK DFLPNEAELMAHFGVSRPTLREAVRVLESERLVEVRRGSRTGARVRVP-G
Mb0505|M.bovis_AF2122/97 STLPPERDLAERLGVNRTSLRQGLARLQQMGLIEVRHG--SGSVVRDPEG
Rv0494|M.tuberculosis_H37Rv STLPPERDLAERLGVNRTSLRQGLARLQQMGLIEVRHG--SGSVVRDPEG
MMAR_4029|M.marinum_M KYIRLD-QLAAELGISVTPVREALFALRAEGLVAQQPHR-----------
MUL_3894|M.ulcerans_Agy99 KYIRLD-QLAAELGISVTPVREALFALRAEGLVAQQPHR-----------
MSMEG_2173|M.smegmatis_MC2_155 DRLPAERDLAAKLGVSRASLSQALVALEVQGVLSVRHGD-GAILIRRP--
Mvan_0294|M.vanbaalenii_PYR-1 TRLPSERELAASMSVSRSSLREAMHELESKRLVSRTRGR--GTIVLAP--
MAB_4644c|M.abscessus_ATCC_199 AKLPGEVALCEEFGVSRATVREAIRDLKTKGLVSTRQGAGVFVTSDRP--
: : * :.:. ..: :.. *. ::
MAV_2560|M.avium_104 RETLARYAGLILEYEGVTVKDVYDARVTLEVPMVEQLAKDRNPKVIAELE
TH_2873|M.thermoresistible__bu RETLARYAGLILEYEGVTVKDVYEARVALEVPMVVQLAKDRNPAVIAELE
Mflv_2114|M.gilvum_PYR-GCK PEIVARPAGLLLELSGATIADLLTAKSAIEPMAARLLAESGSAAAFDELE
Mb0505|M.bovis_AF2122/97 LTHPAVVEALVRKLGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALC
Rv0494|M.tuberculosis_H37Rv LTHPAVVEALVRKLGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALC
MMAR_4029|M.marinum_M -------GFVVLPLTKRDLADVAYVQAHVGGELAARAAINIGAAQLAELQ
MUL_3894|M.ulcerans_Agy99 -------GFVVLPLTKRDLADVAYVQAHVGGELAARAAINIGAAQLAELQ
MSMEG_2173|M.smegmatis_MC2_155 ---TEEGSIRALREHADRIPDIIEAREALEVKLAELAAARRTDAEMAAID
Mvan_0294|M.vanbaalenii_PYR-1 --TASVDELLAIAGGGTEQEHAAELRSVIEPSIAELAAHRATSANVLQLR
MAB_4644c|M.abscessus_ATCC_199 ------LGQWDRVLAAAEIDAVVEIRRLLEVQAAGLAAERRNRSDVAAIR
: : * :
MAV_2560|M.avium_104 QIVERESQLNPGS--EAVDQLT-DFHAAIARLSGNSTLQIVSDMLHHIIE
TH_2873|M.thermoresistible__bu RIVEAESKLEPGA--ETVSQLT-DFHGAIARLSGNKTLQIVTDMLHHVIE
Mflv_2114|M.gilvum_PYR-GCK QLLEELVADSHQS--PRLAEATGAFHLRVVQLSGNATLSIVAGMLHEITV
Mb0505|M.bovis_AF2122/97 AALEVVQQADTAA--ARQAADL-AYFRVLIHSTRNRALGLLYRWVEHAFG
Rv0494|M.tuberculosis_H37Rv AALEVVQQADTAA--ARQAADL-AYFRVLIHSTRNRALGLLYRWVEHAFG
MMAR_4029|M.marinum_M QIQAQLEEAYAADDDERTVRLNHEFHRAINLAADS---PRLSQLMSQITR
MUL_3894|M.ulcerans_Agy99 QIQAQLEEAYAADDDERTVRLNHEFHRAINLAADS---PRLSQLMSQITR
MSMEG_2173|M.smegmatis_MC2_155 AAIATMEKEVEAG--ERGVMGDEMFHEAITSAAHSSLLAKLMHEIAGLIR
Mvan_0294|M.vanbaalenii_PYR-1 DVLDKSHENLLQA---ESLRLDIEFHLLLAQAAQNPLLTALQTLASEWTG
MAB_4644c|M.abscessus_ATCC_199 RALVARAKAESAGDDIEYVDADLEFHRAVVAAVHNPLLDELFDSFSVRMR
:. : . :
MAV_2560|M.avium_104 KANRSLQPTKGTRAEQAVRRSAKTHRMVLDMIKEGDAEKAGELWKRHLQK
TH_2873|M.thermoresistible__bu KANRSLQPTEGTRAEQAVKKSTKTHRMVLDYIKAGDAEKAGELWQKHLQK
Mflv_2114|M.gilvum_PYR-GCK RHTAFVFNERRPVSKADYETLLRSYRRLLQFLRSRDADGAEAHWRRHLDK
Mb0505|M.bovis_AF2122/97 GREHALTG-----AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNA
Rv0494|M.tuberculosis_H37Rv GREHALTG-----AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNA
MMAR_4029|M.marinum_M YQPESVFPG----IEHWPARAVKDHRRVLVALKKHDDKAARTAMSEHLAS
MUL_3894|M.ulcerans_Agy99 YQPESVFPG----IEHWPARAVKDHGQVLVALKKHDDKAARTAMSEHLAS
MSMEG_2173|M.smegmatis_MC2_155 ETRIESLSQ-----ENRPRASLEGHRRIADAIRKQDSQAAAQAMAEHIRM
Mvan_0294|M.vanbaalenii_PYR-1 EVRKHSHST-----RHGRCASVRGHQEIFDAVRTHDGLAARQAMEDHLRD
MAB_4644c|M.abscessus_ATCC_199 EAMLALLAG-----SAERPHDQEHHQWLFDAIVDGSTRRAVELATDQLQH
: : . * :
MAV_2560|M.avium_104 AEEFVLTGAELSTVVDLLE------------
TH_2873|M.thermoresistible__bu AGEYLLSGTEMSTVVDLLE------------
Mflv_2114|M.gilvum_PYR-GCK TRELLLAGLESVPVRDVMG------------
Mb0505|M.bovis_AF2122/97 SALRMVKSYRDRA------------------
Rv0494|M.tuberculosis_H37Rv SALRMVKSYRDRA------------------
MMAR_4029|M.marinum_M SAVPLIDHLIARGVVAESAPSGSAAERARGG
MUL_3894|M.ulcerans_Agy99 SAVPLIDHLIARGVVAESAPSGSAAERARGG
MSMEG_2173|M.smegmatis_MC2_155 VSDVALLREG---------------------
Mvan_0294|M.vanbaalenii_PYR-1 VSELVAKATRS--------------------
MAB_4644c|M.abscessus_ATCC_199 VRMR---------------------------