For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSQRVAIVIGGASGIGWAGAKALAADGCRIVVADRDADAAAARAAELGAPHTSGHVDVTDEESVERLFAE VSTAGPIDIVINCAGFSNVGLITDMPAEDFRSVIDVCLTGGFIVAKHAGRHLPRGGVLVSISSLNARQPA AGMSAYCAAKAGLSMLTQVAALEMGEAGIRVNAIAPGFVHTPLTAPAAMIPGVVEEYVENTALGRAGTPE DIAAAVVFVCSPQASWLTGEVLDLNGGAHLRRYPDVLGHVMRLAQG
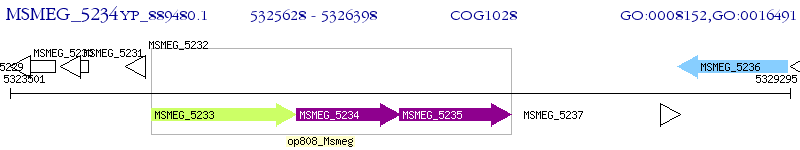
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5234 | - | - | 100% (256) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_5885 | - | 2e-39 | 38.28% (256) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2206 | fabG | 7e-35 | 35.10% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3515c | - | 2e-36 | 36.40% (250) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2071 | - | 1e-112 | 76.77% (254) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3485c | - | 2e-36 | 36.40% (250) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-29 | 35.44% (237) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 1e-31 | 37.18% (234) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4033 | fabG | 3e-34 | 35.39% (243) | 3-oxoacyl- |
| M. avium 104 | MAV_5088 | fabG | 3e-37 | 39.92% (243) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3206 | - | 6e-86 | 79.69% (192) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4042 | - | 1e-34 | 36.11% (252) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4643 | - | 1e-109 | 75.40% (252) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2071|M.gilvum_PYR-GCK ------------------------------------MSNSSE---RVAVI
Mvan_4643|M.vanbaalenii_PYR-1 -----------------------MERVPHHRRKANRMSDSSDRAGREAVV
TH_3206|M.thermoresistible__bu --------------------------------------------------
MSMEG_5234|M.smegmatis_MC2_155 ------------------------------------MS------QRVAIV
MMAR_4033|M.marinum_M -----------------------------------MS--VSLLNGQIAVV
MAV_5088|M.avium_104 -----------------------------------MA--ESLLLGKVAVV
MAB_4680c|M.abscessus_ATCC_199 -----------------------------------MNGNSALLQGKNVIV
MLBr_01807|M.leprae_Br4923 ------------------------MTDAAAKVSAAPGRGKPAFVSRSILV
Mb3515c|M.bovis_AF2122/97 MNSRAPRNLAVSSPSAQVTGRMVQNGENLFQFRREGPQVQLSFQDRTYLV
Rv3485c|M.tuberculosis_H37Rv MNSRAPRNLAVSSPSAQVTGRMVQNGENLFQFRREGPQVQLSFQDRTYLV
MUL_4042|M.ulcerans_Agy99 --------------------------------------MQLSFQDRTYLV
Mflv_2071|M.gilvum_PYR-GCK VGGASGIGWATAQALAAEGCRVTIADRNA---DGARERASELG---SGHR
Mvan_4643|M.vanbaalenii_PYR-1 VGGASGIGWATATALAAEGCRVTIADLNV---EGARERAAGLG---AEHA
TH_3206|M.thermoresistible__bu --------------------------------------------------
MSMEG_5234|M.smegmatis_MC2_155 IGGASGIGWAGAKALAADGCRIVVADRDA---DAAAARAAELG---APHT
MMAR_4033|M.marinum_M TGGAQGLGLAIAERFVSEGARVVLGDVNL---EATEAAAKQLG-GGEVAV
MAV_5088|M.avium_104 TGAGQGIGREIARALHRHGARVVLADLDG---SAARSAAAQIDDSGANCT
MAB_4680c|M.abscessus_ATCC_199 TGGARGLGAAFSRHIVSQGGRVVIADVLD---GDGRQLAAELG---DAAR
MLBr_01807|M.leprae_Br4923 TGGNRGIGLAVARRLVADGHRVAVTHRAS---VVP-----------DWFF
Mb3515c|M.bovis_AF2122/97 TGGGSGIGKGVAAGLVAAGAAVMIVGRNPDKLAAAVKDIEALKT--GAIG
Rv3485c|M.tuberculosis_H37Rv TGGGSGIGKGVAAGLVAAGAAVMIVGRNPDKLAAAVKDIEALKT--GAIG
MUL_4042|M.ulcerans_Agy99 TGGGSGIGKGVAAGLVAAGASVMIVGRNPDKLAGAVQELEALGANGGAIR
Mflv_2071|M.gilvum_PYR-GCK AEAVDVTDEDSVARLFDAI----GPLDIAVNCAG-FSNVGLITDMAAEDF
Mvan_4643|M.vanbaalenii_PYR-1 AAHVDVTDEDSVQRLFEDA----GPIDIAVNCAG-FSNVGLITDMTAADF
TH_3206|M.thermoresistible__bu -----------VARLFDSV----GELDVAVNCAG-FSNVGLITDMPVDQF
MSMEG_5234|M.smegmatis_MC2_155 SGHVDVTDEESVERLFAEVST-AGPIDIVINCAG-FSNVGLITDMPAEDF
MMAR_4033|M.marinum_M AVRCDVTKADEVETLLQTALERFGGLDIMVNNAG-ITRDATMRKMTEEQF
MAV_5088|M.avium_104 GLACDVTSEEQVGALVAGTVREHGGLDVFVNNAG-ITRDASLKRMTVADF
MAB_4680c|M.abscessus_ATCC_199 FTHLDVTDAEQWRELIDTTLAGFGTITGLVNNAG-ISSAAMVADEPLEHF
MLBr_01807|M.leprae_Br4923 GVECDVTDNDAIGAAFKAVEEHQGPVEVLVANAG-ITSDMLIMRMSEEQF
Mb3515c|M.bovis_AF2122/97 YEPADITDEEQTLRVVDAATAWHGRLHGVVHCAGGSQTIGPITQIDSQAW
Rv3485c|M.tuberculosis_H37Rv YEPADITDEEQTLRVVDAATAWHGRLHGVVHCAGGSQTIGPITQIDSQAW
MUL_4042|M.ulcerans_Agy99 YEPTDITNEDETARAVDAVTAWHGRLHGVVHCAGGSENIGPITQVDSEAW
. * : : ** : :
Mflv_2071|M.gilvum_PYR-GCK RAVIDVCLNGAFIVTKHAGRHLRRG--GSLVQISSLNGRQPAAGMSAYCA
Mvan_4643|M.vanbaalenii_PYR-1 RAVIDVCLTGAFIVTKHAGRHLREG--GSLVQISSLNGRQPAAGMSAYCA
TH_3206|M.thermoresistible__bu RSVIDVCLTGAFIVTKHAGRQLREG--GSLVSISSLNARQAAAGMSAYCA
MSMEG_5234|M.smegmatis_MC2_155 RSVIDVCLTGGFIVAKHAGRHLPRG--GVLVSISSLNARQPAAGMSAYCA
MMAR_4033|M.marinum_M DQVIDVHLKGTWNGIRLAAAIMRENKRGAIVNMSSVSGKVGMVGQTNYSA
MAV_5088|M.avium_104 DAVIAVHLRGTWLGVREAAAVMRERKTGSIVNMSSLSGKAGNPGQTNYSA
MAB_4680c|M.abscessus_ATCC_199 RKVIEINLVGVFIGMQAVIPAMRAHGCGSIVNISSAAGLVGLALTSGYGA
MLBr_01807|M.leprae_Br4923 QKVIDVNLTGVFRVAKLASRNMIKQRFGRYIFMGSVVGLSGVPGQANYAA
Mb3515c|M.bovis_AF2122/97 RRTVDLNVNGTMYVLKHAARELVRGGGGSFVGISSIAASNTHRWFGAYGV
Rv3485c|M.tuberculosis_H37Rv RRTVDLNVNGTMYVLKHAARELVRGGGGSFVGISSIAASNTHRWFGAYGV
MUL_4042|M.ulcerans_Agy99 RRTVDLNVNGTMYVLKHAAREMVRGGGGSFVGISSIAASNTHRWFGAYGV
.: : : * : . : * : :.* . * .
Mflv_2071|M.gilvum_PYR-GCK AKAGLSMLTQVAALEMGPRGIRVNAVAPGFVHTPLTAAAASVPGVVEDYV
Mvan_4643|M.vanbaalenii_PYR-1 AKAGLSMLTQVAALELGPRGIRVNAVAPGFVHTPLTAAAASVPGVVEDYV
TH_3206|M.thermoresistible__bu AKAGLSMLTQVAALELAPRGVRVNAVAPGFVHTPLTAPAAAVPGVVEEYL
MSMEG_5234|M.smegmatis_MC2_155 AKAGLSMLTQVAALEMGEAGIRVNAIAPGFVHTPLTAPAAMIPGVVEEYV
MMAR_4033|M.marinum_M AKAGIVGMTKAAAKELAHVGVRVNAIAPGLIRSAMTEAMPQR--IWDQKL
MAV_5088|M.avium_104 AKAGIVGLTKAAAKELAHHNVRVNAIQPGLIRTPMTAAMPPD--VFAERE
MAB_4680c|M.abscessus_ATCC_199 SKWGVRGLSKVAAIELGRERIRVNSVHPGVIYTPMTAKEGFR--EGEGNM
MLBr_01807|M.leprae_Br4923 SKSGLIGMARSLARELGSRSITANVVAPGFIDTEMTRAMDSK--YQEAAL
Mb3515c|M.bovis_AF2122/97 TKSAVDHMMKLAADELGPSWVRVNSIRPGLIRTDLVVPVTESPELSADYR
Rv3485c|M.tuberculosis_H37Rv TKSAVDHMMKLAADELGPSWVRVNSIRPGLIRTDLVVPVTESPELSADYR
MUL_4042|M.ulcerans_Agy99 TKSAVDHLMQLAADELGASWVRVNSIRPGLIRTDLVAAITESAELSSDYA
:* .: : : * *:. : .* : **.: : :.
Mflv_2071|M.gilvum_PYR-GCK DNTALGRAGTPEDIAAAVVFLCSPGTSWMTGEVLDINGGAHLKRYPDVMG
Mvan_4643|M.vanbaalenii_PYR-1 DNTVLGRAGTAEDIANAVVYLCAPGSSWLTGEVLDINGGAHLKRYPDVMG
TH_3206|M.thermoresistible__bu ENTPLGRAGQPEDVAAAVVFLCSPAASWLTGEVLDLNGGAHLKRYPDILG
MSMEG_5234|M.smegmatis_MC2_155 ENTALGRAGTPEDIAAAVVFVCSPQASWLTGEVLDLNGGAHLRRYPDVLG
MMAR_4033|M.marinum_M AEVPMGRAGEPSEVASVALFLACDLSSYMTGTVLDVTGGRFI--------
MAV_5088|M.avium_104 AAVPMKRAGEPEEVAGAVVFLASELSSYITGTVLGVGGGRYM--------
MAB_4680c|M.abscessus_ATCC_199 PIAAMGRVGNADEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPTLTTIT
MLBr_01807|M.leprae_Br4923 QAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH------
Mb3515c|M.bovis_AF2122/97 VCTPLPRVGEVEDVANLAMFLLSDAASWITGQVINVDGGHMLRRGPDFSP
Rv3485c|M.tuberculosis_H37Rv VCTPLPRVGEVEDVANLAMFLLSDAASWITGQVINVDGGHMLRRGPDFSP
MUL_4042|M.ulcerans_Agy99 MCTPLPRQGEVEDVANMAMFLLSDAASFVTGQVINVDGGQMLRRGPDFSA
: * * .::* . :: . :.:::* : : **
Mflv_2071|M.gilvum_PYR-GCK HVMKLMEPAS------
Mvan_4643|M.vanbaalenii_PYR-1 HVMRLVEQSS------
TH_3206|M.thermoresistible__bu HVMKLAEQS-------
MSMEG_5234|M.smegmatis_MC2_155 HVMRLAQG--------
MMAR_4033|M.marinum_M ----------------
MAV_5088|M.avium_104 ----------------
MAB_4680c|M.abscessus_ATCC_199 GQ--------------
MLBr_01807|M.leprae_Br4923 ----------------
Mb3515c|M.bovis_AF2122/97 MLEPVFGADGLRGVVG
Rv3485c|M.tuberculosis_H37Rv MLEPVFGADGLRGVVG
MUL_4042|M.ulcerans_Agy99 MLEPVFGRDALRGLV-