For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IAVAVPEELVDAALRAAAELGRGVADVPAEVIARHAGMSRSTLLRRLGGSRSALDQAVRARGTDPGGLPP VRVRAVGAAAELIGEIGLGAVTLEAIADRAECSVPSLHAVFGTRDGLLREVFEQYSPVRDVEAFLLEDHG DLRDTVRAFYGMLTRALDRPPRVVPAIFSEALARPQSPAVQAVVGHGAPRMLGLLAEWLTGEVAAGRVRD LPVLLLAQQLMAPMLMHLFVRPTSANLAFVGLPDIDSACDVFADTFVRAVTTGESE*
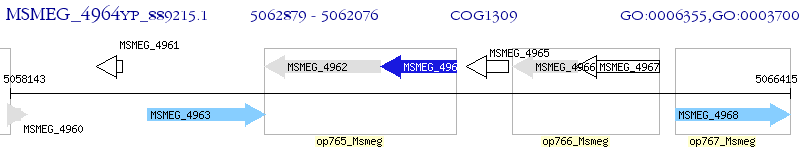
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4964 | - | - | 100% (267) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6503 | - | 8e-06 | 34.55% (110) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0092 | - | 8e-06 | 40.62% (64) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0744 | - | 1e-05 | 32.99% (97) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4757 | - | 7e-73 | 52.69% (260) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 1e-05 | 27.33% (172) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 2e-07 | 28.16% (174) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2910 | - | 4e-05 | 27.22% (158) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4780 | - | 2e-05 | 27.33% (172) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1821 | - | 2e-06 | 26.34% (186) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0340|M.marinum_M -------------------------------MVAD------------FGR
MUL_4780|M.ulcerans_Agy99 -------------------------------MVAD------------FGR
MAV_5165|M.avium_104 ---------------------------------MD------------FGR
Mvan_1821|M.vanbaalenii_PYR-1 -------------------------------MTADRRGDGKTPARAAPGR
MSMEG_4964|M.smegmatis_MC2_155 MIAVAVPEELVDAALRAAAELGRGVADVPAEVIARHAGMSRSTLLRRLGG
MAB_4757|M.abscessus_ATCC_1997 -----MTEDLVNAALQAAHALGKDVADVPLVEVARAAGVSRSTLLRRLGG
TH_2910|M.thermoresistible__bu ------------------------MAAVDPAEPSARGSDDAVDDDDVDVD
Mflv_0744|M.gilvum_PYR-GCK -----------------------MWHNFNVPYTASRG----------PGR
MMAR_0340|M.marinum_M PR----------------DPRIDGAVLRATVELLAETGYAGLLVSAIAKR
MUL_4780|M.ulcerans_Agy99 PR----------------DPRIDGAVLRATVELLAETGYAGLLVSAIAKR
MAV_5165|M.avium_104 PR----------------DPRIDAAVLRATVELLAETGYPGLLVSAIAQR
Mvan_1821|M.vanbaalenii_PYR-1 PR----------------DGRIDAAIIAATRELILQTGYSALSLSAIAAR
MSMEG_4964|M.smegmatis_MC2_155 SRSALDQAVRARGTDPGGLPPVRVRAVGAAAELIGEIGLGAVTLEAIADR
MAB_4757|M.abscessus_ATCC_1997 TRQALDAAVRETGVDPGGRAPVRERATVAAAELIDERGLAAVTLEAVATQ
TH_2910|M.thermoresistible__bu PR----------------KLRSRARLLDAATSLLTSGGVEAVTVDAVTRL
Mflv_0744|M.gilvum_PYR-GCK PP-------------AAKAAETRERILRAAREVFSELGYDAATFQAIAIR
. *: .:: . * . ..*::
MMAR_0340|M.marinum_M AGTSKPAIYRRWPSKAHLVHEAVFPI-DAATAVP-DTGSPVDDLREMVRR
MUL_4780|M.ulcerans_Agy99 AGTSKPAIYRRWPSKAHLVHEAVFPI-DAATAVP-DTGSPVDDLREMVRR
MAV_5165|M.avium_104 AGTSKPAIYRRWPSKAHLVHEAVFPV-GADTAIP-DTGSTADDLREMVRR
Mvan_1821|M.vanbaalenii_PYR-1 AGTTTAAIYRRWSGKVHLVHEAVLS--DAGISVPDDSGDIRGDIRALVET
MSMEG_4964|M.smegmatis_MC2_155 AECSVPSLHAVFGTRDGLLREVFEQY-SPVRDVEAFLLEDHGDLRDTVRA
MAB_4757|M.abscessus_ATCC_1997 ADCSVHSLYAAFGGRDELLRATFDRF-GPIVDIEDTVGDSSVGTEEKLHR
TH_2910|M.thermoresistible__bu SKVARTTLYRHFESTTHLLAEAFERL-LPQVTSPPATGDLRDQLIELVHR
Mflv_0744|M.gilvum_PYR-GCK ADLTRPAINHYFASKGVLWAEVVEQTNATIVSEAIARAHEHTSLIERLSA
: : :: : * .. . :
MMAR_0340|M.marinum_M TTAVLSTPAAK--AALPGLIGEMAADP--TLHSALLVRFGEIFGHGLTTW
MUL_4780|M.ulcerans_Agy99 TTAVLSTPAAK--AAQPGLIGEMAADP--TLHSALLVRFGEIFGHGLTTW
MAV_5165|M.avium_104 TMVFLTTPAAK--AALPGLIGEMAADP--SLHSALLERFAGAIGGGLADW
Mvan_1821|M.vanbaalenii_PYR-1 VRAMFDRPEVR--VALPGLIADTVADP--QLHSMMIARLAG----NLAEF
MSMEG_4964|M.smegmatis_MC2_155 FYGMLTRALDRPPRVVPAIFSEALARPQSPAVQAVVGHGAPRMLGLLAEW
MAB_4757|M.abscessus_ATCC_1997 IYQRLVQAFSQKPRVMPAMYAEIMARPFDPSVRKLIEHNAPRMLGSVGLW
TH_2910|M.thermoresistible__bu QAALIDEAPLHLTALAWLALGPDPNSDARQPLAGLSARVIEQYRQPFDEL
Mflv_0744|M.gilvum_PYR-GCK FLTVATQAESDDRSAAAFLVTSVLESQRHPELINDEHDSLKNSRRFVSAA
. .
MMAR_0340|M.marinum_M LDNAAARGQVRADVTAPELAEAIAGIVL-IGLLTR-PGELEPGVGDAWID
MUL_4780|M.ulcerans_Agy99 LDNAAARGQVRADVTAPELAEAIAGIVL-IGLLTR-PGELEPGVGDAWID
MAV_5165|M.avium_104 LAAAAARGEARPDVTAAELAETIAGVTL-VALLTR-PTELD----DAWVD
Mvan_1821|M.vanbaalenii_PYR-1 ETRFGQ--QRRSDDELPMLAEIVAGSAI-FRILVRRDAALD----DAWVD
MSMEG_4964|M.smegmatis_MC2_155 LTGEVAAGRVR-DLPVLLLAQQLMAPML-MHLFVRPTSANLAFVGLPDID
MAB_4757|M.abscessus_ATCC_1997 LSGEIAAGRIR-DLPVTVLTQQLLAPVV-MHTALRPAAEGVLGLELPDIQ
TH_2910|M.thermoresistible__bu LAGAAARDELD-DFDTTLAIIQLLGPIV-FAKLTGMKTLTP----ADRVR
Mflv_0744|M.gilvum_PYR-GCK VSDAIERGELSADTDVTHLVEMLVAVVWGMGFYAGFVGDRS---DLSAVV
. * : . . :
MMAR_0340|M.marinum_M RTTTLLLKGIRKGTPQ------
MUL_4780|M.ulcerans_Agy99 RTTTLLLKGIRKGTPQ------
MAV_5165|M.avium_104 RTTRLLLKGISA----------
Mvan_1821|M.vanbaalenii_PYR-1 AVTEMITERWPAG---------
MSMEG_4964|M.smegmatis_MC2_155 SACDVFADTFVRAVTTGESE--
MAB_4757|M.abscessus_ATCC_1997 EVCKIFADAFLHGVRVPEPPRG
TH_2910|M.thermoresistible__bu IVDDFLVAHRPPTPETA-----
Mflv_0744|M.gilvum_PYR-GCK HKLELLLAGKLWNVAD------
.: