For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVSTDRPAARESEHTGIARWIRRLAIPIILGWIALLGVLGATVPSLEEVGQMRSVSMSPDYAPSVIAMK RVGEVFEEFKSDSSAMIVLEGDEPLGDDAHIFYNDMVDRLEADTEHVEHVQDFWGDPLTEAGAQSNDGRA AYVQVFLAGNMGEALANESVQAVQELVEGLQPPPGVKVYVTGGSALANDQQISGDRSVRIIEAVTFLVII TMLLLVYRSIVTVLITLFLVVICLQSARGVVAFLGYHELIGLSTFATQLLVTLAIAASTDYAIFLIGRYQ EARTLGESKEQAYYTMFRGTAHVVLGSGMTIAGATLCLHFTRLPYFQTLGIPMAVGMTIAVVAALTLGPA VITVMTRFGKILEPKRAMRIRGWRRLGAFVVRWPGAVLMATVLLSLVGLLALPGYRTNYNDRNYLPDDLP AQEGFAAAERHFSPAKMNPELLLIETDHDIRNSADFLVIDKIAKAVFRVPGIGSVQAITRPQGEPLEFST IPAQMSMGGTLQTMNRKYMLDRADDMLLQADEMQRTIELMGRTIELMEEMAATTHSMVGKTHLMVEDVKE LRDNISNFDDFLRPLRNYLYWEPHCYDIPMCHSMRSIFDALDGMDTMTEAMEELLPDMDRLDALMPRMVA LMPPQIETMRKMRTMMLTMHQTQKGLQEQMGAMQEGQTAMGEAFNTAKNDDTFYLPPEIFNNADFKRGMD SFISPDGKAVRFIISHEGDPLTPEGIKLIDQIKLAAKEAMKNTPWEGSKIYVGGTAAAFKDMQEGNNYDL IIAGISALCLIFIIMLLITRSVVAAAVIVGTVLVSLGASFGLSILVWQHILGIDLHFMVMAMAVIVLLAV GADYNLLLVARLKEELPAGINTGIIRAMGGSGSVVTAAGLVFAFTMMSMLVSEMTVVAQVGTTIGMGLLF DTLVIRSFMTPSIAALLGRWFWWPQVVRARPARGVVARALERDKAATH
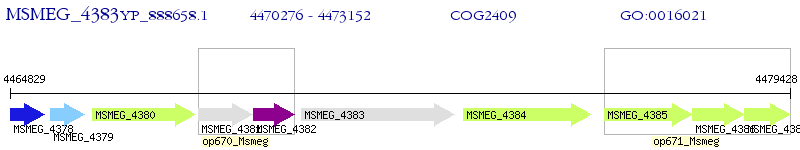
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4383 | - | - | 100% (958) | MmpL5 protein |
| M. smegmatis MC2 155 | MSMEG_0225 | - | 0.0 | 72.43% (943) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_3496 | - | 0.0 | 72.04% (937) | MmpL4 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0695c | mmpL5 | 0.0 | 69.57% (940) | transmembrane transport protein MmpL5 |
| M. gilvum PYR-GCK | Mflv_3435 | - | 0.0 | 71.63% (934) | transport protein |
| M. tuberculosis H37Rv | Rv0676c | mmpL5 | 0.0 | 69.47% (940) | transmembrane transport protein MmpL5 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 61.08% (930) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_2301 | - | 0.0 | 63.33% (938) | putative membrane protein, mmpL |
| M. marinum M | MMAR_1005 | mmpL5 | 0.0 | 69.30% (935) | transmembrane transport protein, MmpL5 |
| M. avium 104 | MAV_2510 | - | 0.0 | 68.63% (934) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 65.92% (942) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_0757 | mmpL5 | 0.0 | 69.09% (935) | transmembrane transport protein, MmpL5 |
| M. vanbaalenii PYR-1 | Mvan_3189 | - | 0.0 | 71.84% (934) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3435|M.gilvum_PYR-GCK MSAPVSDARTDEFPAAHGPHRS------FIPRMIRLFAVPIILGWIALIV
Mvan_3189|M.vanbaalenii_PYR-1 MSAPVSDARTDEFPAARQPHRP------FIPRMIRLFAVPIILGWIALIV
TH_3260|M.thermoresistible__bu MTANPGHVRTDEAPTDRLDIKVSTHGPSFISTWIRRLSLPIIVGWIALIA
Mb0695c|M.bovis_AF2122/97 MIVQRTAAPTGSVPPDRHAARP------FIPRMIRTFAVPIILGWLVTIA
Rv0676c|M.tuberculosis_H37Rv MIVQRTAAPTGSVPPDRHAARP------FIPRMIRTFAVPIILGWLVTIA
MMAR_1005|M.marinum_M MTATTTDTPTDVVPPAEHPPRP------FIPRMIRTFAVPLILGWVLLIA
MUL_0757|M.ulcerans_Agy99 MTATTTDTPTDVVPPAEHPPRP------FIPRMIRTFAVPLILGWVLLIA
MAV_2510|M.avium_104 MSTTFDDTRTDAIPVVKEPVRE------KIPRFIRAFAVPIIIGWIALIA
MSMEG_4383|M.smegmatis_MC2_155 ------MAVSTDRPAARESEHTG------IARWIRRLAIPIILGWIALLG
MLBr_02378|M.leprae_Br4923 MTVKCANDLDTHTKPP------------FVARMIHRFAVPIILVWLAIAV
MAB_2301|M.abscessus_ATCC_1997 MSGSHAG--RSAAEPT------------FLAKWIRRLSIPIVIGWLAIVF
:. *: :::*::: *:
Mflv_3435|M.gilvum_PYR-GCK VVNVTVPQLETVGEAQAVSMSPNDAPSMISMKKVGELFEEGDSDSSVMIV
Mvan_3189|M.vanbaalenii_PYR-1 ILNVTVPQLEAVGEARAVSMSPNEAPSLISMKKVGELFREGDSDSSVMIV
TH_3260|M.thermoresistible__bu LLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGELFDEGSSDSQVMIV
Mb0695c|M.bovis_AF2122/97 VLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAMIV
Rv0676c|M.tuberculosis_H37Rv VLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAMIV
MMAR_1005|M.marinum_M VLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAMIV
MUL_0757|M.ulcerans_Agy99 VLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAMIV
MAV_2510|M.avium_104 VLNVVVPQLDEVGKMRSVEMTPDDAQSVVATKRMGAVFNEYKSNSSVMIV
MSMEG_4383|M.smegmatis_MC2_155 VLGATVPSLEEVGQMRSVSMSPDYAPSVIAMKRVGEVFEEFKSDSSAMIV
MLBr_02378|M.leprae_Br4923 TVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVFNEGNSDSVIMIV
MAB_2301|M.abscessus_ATCC_1997 ALNTFVPQLEQVGKEHTVSLSPQSAPSMQAMKHMGKMFKESDSDSVVMIV
:. :*.*: **: ::*.::*. * * : :::* :* * .*:* ***
Mflv_3435|M.gilvum_PYR-GCK FEGDQPLGDEAHAWYDELVDRLEADTAHVQSVQDFWSDPLTASGSQSNDG
Mvan_3189|M.vanbaalenii_PYR-1 FEGDQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDPLTASGSQSNDG
TH_3260|M.thermoresistible__bu LEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWSDPLTATGVQSPDG
Mb0695c|M.bovis_AF2122/97 LEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSSDG
Rv0676c|M.tuberculosis_H37Rv LEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSSDG
MMAR_1005|M.marinum_M LEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSGDG
MUL_0757|M.ulcerans_Agy99 LEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSGDG
MAV_2510|M.avium_104 LEGQQPLGADAHAYYDEIVRRLNADTKHVEHVQDMWSDPLTGAGAQSNDG
MSMEG_4383|M.smegmatis_MC2_155 LEGDEPLGDDAHIFYNDMVDRLEADTEHVEHVQDFWGDPLTEAGAQSNDG
MLBr_02378|M.leprae_Br4923 LEGDKPLGDDAHRFYDGLIRKLRAD-KHVQSVQDFWGDPLTAPGAQSNDG
MAB_2301|M.abscessus_ATCC_1997 LEGQERLGPGSHKFYDEMVARLRADTKHVQHVQDFWGDPLTEAGAQSTDG
:**:. ** :: :*: :: :*.** **: :**:*.**** .* ** **
Mflv_3435|M.gilvum_PYR-GCK KAAYVQVKLSGNQGEALANESVKAVQQTVSSLE---------PPPGVRAF
Mvan_3189|M.vanbaalenii_PYR-1 KAAYVQVKLAGNQGESLANESVQAAQEIVRSLE---------PPPGVRAF
TH_3260|M.thermoresistible__bu KSAYVQVKLAGNQGEALANESVETVREMIATVSEE------MAPEGVQAF
Mb0695c|M.bovis_AF2122/97 KAAYVQVKLAGNQGESLANESVEAVKTIVERLA---------PPPGVKVY
Rv0676c|M.tuberculosis_H37Rv KAAYVQVKLAGNQGESLANESVEAVKTIVERLA---------PPPGVKVY
MMAR_1005|M.marinum_M KAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---------APPGVKVY
MUL_0757|M.ulcerans_Agy99 KAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---------APPGVKVY
MAV_2510|M.avium_104 KASYVQVYLAGNQGEALANESVESVQNIVKSVQ---------APNGVKAY
MSMEG_4383|M.smegmatis_MC2_155 RAAYVQVFLAGNMGEALANESVQAVQELVEGLQ---------PPPGVKVY
MLBr_02378|M.leprae_Br4923 KAAYVQLSLAGNQGELLAQESIDAVRKIVVQTPA---------PPGITAW
MAB_2301|M.abscessus_ATCC_1997 KATYVQVNLAGNMGETLSNESTEAARNIVKQLEADWTKDGNKLPDGLTVY
:::***: *:** ** *::** .:.: : * *: .:
Mflv_3435|M.gilvum_PYR-GCK VTGPAALAADQHIAGDRSMRLIEAATFTVIIVMLLLVYRSIVTVLITLVM
Mvan_3189|M.vanbaalenii_PYR-1 VTGPAALAADQHIASDRSVRVIELVTFAVIIVMLLLVYRSIVTVLLTLVM
TH_3260|M.thermoresistible__bu VTGGAAMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVITVLIALFM
Mb0695c|M.bovis_AF2122/97 VTGSAALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTM
Rv0676c|M.tuberculosis_H37Rv VTGSAALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTM
MMAR_1005|M.marinum_M VTGSAALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTM
MUL_0757|M.ulcerans_Agy99 VTGSAALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTM
MAV_2510|M.avium_104 VTGPAALSADQHTAGDRSLQLITAATFTVIIGMLLLVYRSVITVLLTLVM
MSMEG_4383|M.smegmatis_MC2_155 VTGGSALANDQQISGDRSVRIIEAVTFLVIITMLLLVYRSIVTVLITLFL
MLBr_02378|M.leprae_Br4923 VTGASALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITVILLLFT
MAB_2301|M.abscessus_ATCC_1997 VTGPGALQTDMNHAGDGSLQLITGLTFLVIVVMLLFFYRSIFTVIMVFML
*** .*: * : :.* *: * :. ** **:.***..* : :
Mflv_3435|M.gilvum_PYR-GCK VVMSLATARGVVAFLGHHEIIGLSTFATNLLVTLAIAAATDYAIFLIGRY
Mvan_3189|M.vanbaalenii_PYR-1 VVLSLATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIFLIGRY
TH_3260|M.thermoresistible__bu VLIGLLTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYAIFLIGRY
Mb0695c|M.bovis_AF2122/97 VVLGLLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRY
Rv0676c|M.tuberculosis_H37Rv VVLGLLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRY
MMAR_1005|M.marinum_M VVLGLLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRY
MUL_0757|M.ulcerans_Agy99 VVLGLLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRY
MAV_2510|M.avium_104 VVLELSAARGMVAFLGYYKIIGLSTFATNLLVTLAIAAATDYAIFLIGRY
MSMEG_4383|M.smegmatis_MC2_155 VVICLQSARGVVAFLGYHELIGLSTFATQLLVTLAIAASTDYAIFLIGRY
MLBr_02378|M.leprae_Br4923 VGIESAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGIFITGRY
MAB_2301|M.abscessus_ATCC_1997 VGIQLSAARGVIAFLGNTEIIGLSTFAVNLLVMLSIAAGTDYAIFLIGRY
* : :** :*:** :**** **..**. *:** ..**.**: ***
Mflv_3435|M.gilvum_PYR-GCK QEARTAGEDKETAYYTMFHGTAHVVLGSGLTIAGATFCLSFTRLPYFQTM
Mvan_3189|M.vanbaalenii_PYR-1 QEARTNGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLPYFQTL
TH_3260|M.thermoresistible__bu QEARSVGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTRLPYFQSL
Mb0695c|M.bovis_AF2122/97 QEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTL
Rv0676c|M.tuberculosis_H37Rv QEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTL
MMAR_1005|M.marinum_M QEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTL
MUL_0757|M.ulcerans_Agy99 QEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTL
MAV_2510|M.avium_104 QEARAVGESREDAYYTMYKGTAHVVAGSGMTIAGATFCLHFTNLPYFQTL
MSMEG_4383|M.smegmatis_MC2_155 QEARTLGESKEQAYYTMFRGTAHVVLGSGMTIAGATLCLHFTRLPYFQTL
MLBr_02378|M.leprae_Br4923 QEARQANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARMPYFQTL
MAB_2301|M.abscessus_ATCC_1997 QEARAMGADKEAAYYEMFHGTAHVILGSGMTIAGAMYCLSFTRMPYFQTL
**** . .: *:: *: ** **: .**:**:** ** *:.:****::
Mflv_3435|M.gilvum_PYR-GCK GVPLAIGMFVVVMAGVVLMVACISVATRFGKLLEPKRAMRIRGWRKIGAA
Mvan_3189|M.vanbaalenii_PYR-1 GVPLAIGMFVVVMAGVILMVAMISVATRFGKLLEPKRAMRIRGWRKIGAA
TH_3260|M.thermoresistible__bu GPPLAIGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARVRFWRKVGAS
Mb0695c|M.bovis_AF2122/97 GVPLAIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAA
Rv0676c|M.tuberculosis_H37Rv GVPLAIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAA
MMAR_1005|M.marinum_M GVPLAIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAA
MUL_0757|M.ulcerans_Agy99 GVPLAIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAA
MAV_2510|M.avium_104 GIPLAIGMVVVVAAALTLGPAVISVASRFRQTLEPRRTQRIRGWRKVGTV
MSMEG_4383|M.smegmatis_MC2_155 GIPMAVGMTIAVVAALTLGPAVITVMTRFGKILEPKRAMRIRGWRRLGAF
MLBr_02378|M.leprae_Br4923 GVPCAVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKVRGWRRIGTV
MAB_2301|M.abscessus_ATCC_1997 GVPCSVGLLVGVLVSLTLGPALITIGSRFG-LFEPKRAMRIRSWRKIGTT
* * ::*: : : ..: : * ::: :** :**:* ::* **::*:
Mflv_3435|M.gilvum_PYR-GCK IVRWPGPILVATMAITLVGLLALPGYKTNYNDRDYLPADLPANQGYAVAD
Mvan_3189|M.vanbaalenii_PYR-1 VVRWPGPILVATLAITLVGLLALPGYKTNYNDRTYLPADLPANEGYAVAD
TH_3260|M.thermoresistible__bu VVRWPGPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSNAGYEAAE
Mb0695c|M.bovis_AF2122/97 IVRWPGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAE
Rv0676c|M.tuberculosis_H37Rv IVRWPGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAE
MMAR_1005|M.marinum_M IVRWPGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAE
MUL_0757|M.ulcerans_Agy99 IVRWPGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAE
MAV_2510|M.avium_104 VVRWPGPILVLTIGVALVGLLTLPGYRTNYNDRNYLPTDLPANEGYAAAD
MSMEG_4383|M.smegmatis_MC2_155 VVRWPGAVLMATVLLSLVGLLALPGYRTNYNDRNYLPDDLPAQEGFAAAE
MLBr_02378|M.leprae_Br4923 VVRWPLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQGFAAAD
MAB_2301|M.abscessus_ATCC_1997 IVRWPGPVLAAAMAIAIIGLAALPGYRTSYDDKKYIPKDIPANAGFQAAD
:**** .:: : ::::** :****:*.*.*: *:* .:*:: *: .*:
Mflv_3435|M.gilvum_PYR-GCK EHFDQARMNPELLMIESDHDLRNSADFLVIDKIAKALFRVEGIARVQAIT
Mvan_3189|M.vanbaalenii_PYR-1 RHFDQARMNPELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIARVQAIT
TH_3260|M.thermoresistible__bu RHFDPARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGISRVQGIT
Mb0695c|M.bovis_AF2122/97 RHFSQARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAIT
Rv0676c|M.tuberculosis_H37Rv RHFSQARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAIT
MMAR_1005|M.marinum_M RHFSQARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAIT
MUL_0757|M.ulcerans_Agy99 RHFSQARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAIT
MAV_2510|M.avium_104 RHFSQARMNPEVLMIESDHDLRNSADFLVIDKIAKTVFRVPGIGRVQAIT
MSMEG_4383|M.smegmatis_MC2_155 RHFSPAKMNPELLLIETDHDIRNSADFLVIDKIAKAVFRVPGIGSVQAIT
MLBr_02378|M.leprae_Br4923 RHFPQARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGISRVQAIT
MAB_2301|M.abscessus_ATCC_1997 RHFSQARMNPEMLLIESDHDMRNSADFLVIDKIAKAVFRVPGIARVQAIT
.** *:::**:*::*:***:**.:***:::*:*: :* * **. **.**
Mflv_3435|M.gilvum_PYR-GCK RPDGKPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIKT
Mvan_3189|M.vanbaalenii_PYR-1 RPDGKPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIAT
TH_3260|M.thermoresistible__bu RPEGTPIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADEMQLMIDT
Mb0695c|M.bovis_AF2122/97 RPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQ
Rv0676c|M.tuberculosis_H37Rv RPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQ
MMAR_1005|M.marinum_M RPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQ
MUL_0757|M.ulcerans_Agy99 RPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQ
MAV_2510|M.avium_104 RPQGTPIEHTSIPFQISMQGVTQQMNQKYQQDQMADMLHQADMMQTTIDS
MSMEG_4383|M.smegmatis_MC2_155 RPQGEPLEFSTIPAQMSMGGTLQTMNRKYMLDRADDMLLQADEMQRTIEL
MLBr_02378|M.leprae_Br4923 RPDGTAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENMAETIAS
MAB_2301|M.abscessus_ATCC_1997 RPQGTPIEHSSIPFLISMQSVGQRQNMKLMKDRMADMKVQADKMGENVVI
**:* .:..::** :* .. * . : * : *.: : :
Mflv_3435|M.gilvum_PYR-GCK MEKMSGLTAQMADITHSMVTKMEGMVVDIEDLRDSIANFDDFFRPIRNYF
Mvan_3189|M.vanbaalenii_PYR-1 MEKMSNLTAQMADITHSMVSKMENMLTDIEDLRDSIANFDDFFRPIRNYF
TH_3260|M.thermoresistible__bu MDKMIALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFFRPIRNYF
Mb0695c|M.bovis_AF2122/97 MERMHSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYF
Rv0676c|M.tuberculosis_H37Rv MERMHSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYF
MMAR_1005|M.marinum_M MERMHDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYF
MUL_0757|M.ulcerans_Agy99 MERMHDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYF
MAV_2510|M.avium_104 MEKMQSITVQMSNDMHVMVQKMHDMTIDINDLRNKMADFEDFFRPMRSYF
MSMEG_4383|M.smegmatis_MC2_155 MGRTIELMEEMAATTHSMVGKTHLMVEDVKELRDNISNFDDFLRPLRNYL
MLBr_02378|M.leprae_Br4923 MRRMHQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWRPIRSYF
MAB_2301|M.abscessus_ATCC_1997 MKRTLANMTTMAGITHSLVGDMHTLQGTIHEMRDSISNFDDWFRPIRNYL
* : : . :: . : .:*: :::*:*: **:*.*:
Mflv_3435|M.gilvum_PYR-GCK YWEPHCYNIPVCWALRSVFDTLDGINIMTDDFKAIIPDMKRLDTLMPQMV
Mvan_3189|M.vanbaalenii_PYR-1 YWEPHCYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDSLMPQMV
TH_3260|M.thermoresistible__bu YWEPHCYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLNDLMPQML
Mb0695c|M.bovis_AF2122/97 YWEKHCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLT
Rv0676c|M.tuberculosis_H37Rv YWEKHCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLT
MMAR_1005|M.marinum_M YWEKHCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMT
MUL_0757|M.ulcerans_Agy99 YWEKHCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMT
MAV_2510|M.avium_104 YWEKHCYDIPVCWSLRSVFDALDGIDTMTDDIQSLLPIMDHLDTLMPQMV
MSMEG_4383|M.smegmatis_MC2_155 YWEPHCYDIPMCHSMRSIFDALDGMDTMTEAMEELLPDMDRLDALMPRMV
MLBr_02378|M.leprae_Br4923 YWERHCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDLLMPQML
MAB_2301|M.abscessus_ATCC_1997 YWEKHCFDIPMCWAMRSIFDTLDKIDEMTETMDGMVVNMERMDTLMPKMV
*** **::**:* ::**:**:** :: : : : :: :..:: ***::
Mflv_3435|M.gilvum_PYR-GCK ALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMN
Mvan_3189|M.vanbaalenii_PYR-1 ALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMN
TH_3260|M.thermoresistible__bu TLLPTTVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGEAFDQARN
Mb0695c|M.bovis_AF2122/97 AMMPEMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRN
Rv0676c|M.tuberculosis_H37Rv AMMPEMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRN
MMAR_1005|M.marinum_M AMMPEMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRN
MUL_0757|M.ulcerans_Agy99 AMMPEMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRN
MAV_2510|M.avium_104 ALMPSMIENMKAMKTTMLTMYSTQKGLQDQQNEAQKNSNAMGKAFDASKN
MSMEG_4383|M.smegmatis_MC2_155 ALMPPQIETMRKMRTMMLTMHQTQKGLQEQMGAMQEGQTAMGEAFNTAKN
MLBr_02378|M.leprae_Br4923 EQFPPMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKAFDTAKN
MAB_2301|M.abscessus_ATCC_1997 SDMSAMIPIMEDMQRMMLTQHSTMSGFYNQMDEMSQNSTAMGKAFDAAKN
:. : *. *: :*: : * :* ... :**:**: : *
Mflv_3435|M.gilvum_PYR-GCK DDSFYLPPEIFENADFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKI
Mvan_3189|M.vanbaalenii_PYR-1 DDSFYLPPEIFENKDFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKI
TH_3260|M.thermoresistible__bu DDSFYLPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMTPEGLAKI
Mb0695c|M.bovis_AF2122/97 DDSFYLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARI
Rv0676c|M.tuberculosis_H37Rv DDSFYLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARI
MMAR_1005|M.marinum_M DDSFYLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHI
MUL_0757|M.ulcerans_Agy99 DDSFYLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHI
MAV_2510|M.avium_104 DDSFYLPPETFNNKEFKKGMKNFISPDGHAVRFIISHDGDPMTQEGISHI
MSMEG_4383|M.smegmatis_MC2_155 DDTFYLPPEIFNNADFKRGMDSFISPDGKAVRFIISHEGDPLTPEGIKLI
MLBr_02378|M.leprae_Br4923 DDSFYLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASVAGIASI
MAB_2301|M.abscessus_ATCC_1997 DDSFYLPPEIFDNADFKRGMKSFLSPDGKAVRMIISHRGDPATPEGLSHV
**:****** *.* :*::.:..*:*.**:*.*:** * *** : *: :
Mflv_3435|M.gilvum_PYR-GCK DKIKTAAKEAVKGTPLEGSKIYLGGTAAVFKDMQDGNNYDLLIAGIAALG
Mvan_3189|M.vanbaalenii_PYR-1 DKIKTAAKEAIKGTPLEGSKIYLGGTAATFKDMQDGNNYDLLIAGISALG
TH_3260|M.thermoresistible__bu EPIKKAAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLIIAAIAAVC
Mb0695c|M.bovis_AF2122/97 AKIKTAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALC
Rv0676c|M.tuberculosis_H37Rv AKIKTAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALC
MMAR_1005|M.marinum_M DKIKTAAKEAIKGTPLEGSTIYLGGTAAMFKDLSDGNTFDLMIAGISALC
MUL_0757|M.ulcerans_Agy99 DKIKTAAKEAIKGTPLEGSTIYLGGTVAMFKDLSDGNTFDLMIAGISALC
MAV_2510|M.avium_104 GAIKKAAYEALKGTPLEGSKIYLAGTASIYKDLSDGNNYDLLIAGISSLC
MSMEG_4383|M.smegmatis_MC2_155 DQIKLAAKEAMKNTPWEGSKIYVGGTAAAFKDMQEGNNYDLIIAGISALC
MLBr_02378|M.leprae_Br4923 NAIRTAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIAGISSLC
MAB_2301|M.abscessus_ATCC_1997 EPIKQAAIEAVKGTPLEDAKIELGGTAAVFKDMSDGAKYDLMIAGISALC
*: ** **:*.** *.: : :.**.: :**:.:* .:**:**.*:::
Mflv_3435|M.gilvum_PYR-GCK LIFIIMLILTRAVVAAAVIVGTVVLSLAASFGLSVLFWQHILGQPLHWMV
Mvan_3189|M.vanbaalenii_PYR-1 LIFIIMLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGTELHWMV
TH_3260|M.thermoresistible__bu LIFIIMLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVGLEMHWMV
Mb0695c|M.bovis_AF2122/97 LIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLV
Rv0676c|M.tuberculosis_H37Rv LIFIIMLITTRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLV
MMAR_1005|M.marinum_M LIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLV
MUL_0757|M.ulcerans_Agy99 LIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHVLGIELHWLV
MAV_2510|M.avium_104 LIFIIMLIITRGVVASAVIVGTVLLSLGASFGLSVLIWQHLIGIELHWMV
MSMEG_4383|M.smegmatis_MC2_155 LIFIIMLLITRSVVAAAVIVGTVLVSLGASFGLSILVWQHILGIDLHFMV
MLBr_02378|M.leprae_Br4923 LIFIIMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGIELHYLV
MAB_2301|M.abscessus_ATCC_1997 LIFLIMLLITRSFVASLVIVGTVALSLGASFGLSVIIWQYILGIELHWLV
***:***: **..**: ****** :**.****:*::.**:::* :*::*
Mflv_3435|M.gilvum_PYR-GCK LAMAVIILLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAG
Mvan_3189|M.vanbaalenii_PYR-1 LAMAVIVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAG
TH_3260|M.thermoresistible__bu FPMALIILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSGSVVTAAG
Mb0695c|M.bovis_AF2122/97 LAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAG
Rv0676c|M.tuberculosis_H37Rv LAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAG
MMAR_1005|M.marinum_M LAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAG
MUL_0757|M.ulcerans_Agy99 LAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAG
MAV_2510|M.avium_104 LAMSVIILLAVGADYNLLLVARFKEEIHAGLNTGIIRSMGGTGSVVTSAG
MSMEG_4383|M.smegmatis_MC2_155 MAMAVIVLLAVGADYNLLLVARLKEELPAGINTGIIRAMGGSGSVVTAAG
MLBr_02378|M.leprae_Br4923 LAMSVIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGKVVTNAG
MAB_2301|M.abscessus_ATCC_1997 MQMAIIVLLAVGSDYNLLLVSRLKEELHGGLKTGIIRSMGGTGSVVTSAG
: *::*:*****:*****:*:*:::*: .*: **:**:***:*.*** **
Mflv_3435|M.gilvum_PYR-GCK LVFALTMMSMSVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGP
Mvan_3189|M.vanbaalenii_PYR-1 LVFALTMMSMAVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGP
TH_3260|M.thermoresistible__bu LVFGLTMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPSIATLLGR
Mb0695c|M.bovis_AF2122/97 LVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGK
Rv0676c|M.tuberculosis_H37Rv LVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGK
MMAR_1005|M.marinum_M LVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGK
MUL_0757|M.ulcerans_Agy99 LVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGK
MAV_2510|M.avium_104 LVFAFTMMTMAVSELTVIGQVGTTIGLGLLFDTLIVRSLMTPSIAALLGK
MSMEG_4383|M.smegmatis_MC2_155 LVFAFTMMSMLVSEMTVVAQVGTTIGMGLLFDTLVIRSFMTPSIAALLGR
MLBr_02378|M.leprae_Br4923 LVFAFTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSIAALLGR
MAB_2301|M.abscessus_ATCC_1997 LVFAFTMAVMVVSDLRIIGQVGTTIALGLLFDTLIVRSFMTPAVATLLGR
***.:** : **:: ::.******.:******:::*::* *::*:***
Mflv_3435|M.gilvum_PYR-GCK WFWWPQRVRTRPVPAPWPRSGGLQSDPSEGVRA----------------
Mvan_3189|M.vanbaalenii_PYR-1 WFWWPQRVRTRPVPAPWPRPGGLQSDPSEGVKV----------------
TH_3260|M.thermoresistible__bu WFWWPQFVRKRPVPQPWPSVPPRGPATTDRVSV----------------
Mb0695c|M.bovis_AF2122/97 WFWWPQVVRQRPVPQPWPSPASARTFALV--------------------
Rv0676c|M.tuberculosis_H37Rv WFWWPQVVRQRPIPQPWPSPASARTFALV--------------------
MMAR_1005|M.marinum_M WFWWPQVVRQRPAPQPWPTPAPTP--ATTSV------------------
MUL_0757|M.ulcerans_Agy99 WFWWPQVVRQRPAPQPWPTPAPTPPPATTSV------------------
MAV_2510|M.avium_104 WFWWPQRVRQRPVPSPWPKPAAQEREVLADA------------------
MSMEG_4383|M.smegmatis_MC2_155 WFWWPQVVRARPARGVVARALERDKAATH--------------------
MLBr_02378|M.leprae_Br4923 WFWWPQQGRTRPLLTVAAPVGLPPDRSLVYSD-----------------
MAB_2301|M.abscessus_ATCC_1997 WFWWPLNVRTRPLPPPRTTVAADPTPPPVPVGPGHGDDPITTEFPRPSV
***** * ** .