For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STPDAPTEAIPSAHGKYTNHGGIAKLIRTLAVPIILVWVALIAVLNTIVPQLEEVGEMRSVSMSPDDAPS MIAMKRVGEVFEEFKSNSSVMIVLEGEQPLGDEAHKYYDEIVDKLEADKTHIEHVQDFWGDPLTASGAQS SDGLASYVQVYTAGNQGEALANESVKAVQDIVDSVQAPPGVTAYVTGPAAMSADQQIAGDRSMRMIEALT FTVIIIMLLMVYRSIITVILSLVMVVFSLAAARGVIAFLGYYEIIGLSTFATNLLVTLAIAAATDYAIFL IGRYQEARSVGESREQAYYTMFHSTAHVVLGSGMTIAGATLCLHFTRMPYFQSLGIPLAIGMTVVVLTAL AVGPAIITVASRFGKTLEPKRAMRTRGWRKVGAAVVRWPGPILVAAVGLSMIGLLALPGYRTNYNDRDYQ PADLPANSGYAAADRHFSQARMNPELLMIESDHDLRNSADFLVIERIAKRVVGVPGVSRVQAITRPQGTP IEHTSIPFTISMQGTTQTMNQKYMQDRMADMLVQADQMQVTVDTMTKMIALMEQMRDTMNSMVGKMHGMV DDIKELRDNISDFDDFFRPIRNYLYWEPHCYNIPMCYSLRSIFDTMDGIDTMTADFEQIVPDMDRMNALI PVMIANMEPMITTMKTMKTMMLTMQATNAGLQDQMAAMQENSTAMGEAFDKAKNDDSFYLPPETFDNPDF KRGMKMFLSPDGHAVRFIISHEGDPMSPEGIQKIDDIKLAAKEAIKGTPLEGSKIYLGGTAATFKDMQEG ANYDLIIAGIAALCLIFIIMLIITRAVVASAVIVGTVVISLGSAFGLSVLIWQHLVGLELHWMVLAMAVI VLLAVGADYNLLLVSRIKEEIHAGLDTGMIRAMGGSGSVVTAAGLVFAFTMMSMAVSELAIIGQVGTTIG LGLLFDTLVIRSFMTPSIAALLGKWFWWPQMVRRRPKPSPWPKPIQREPADAVN*
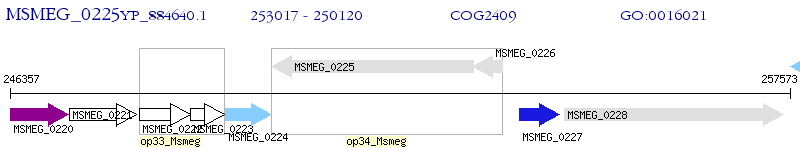
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0225 | - | - | 100% (965) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_3496 | - | 0.0 | 83.45% (967) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_4383 | - | 0.0 | 72.43% (943) | MmpL5 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0695c | mmpL5 | 0.0 | 71.16% (950) | transmembrane transport protein MmpL5 |
| M. gilvum PYR-GCK | Mflv_3435 | - | 0.0 | 73.97% (968) | transport protein |
| M. tuberculosis H37Rv | Rv0676c | mmpL5 | 0.0 | 71.05% (950) | transmembrane transport protein MmpL5 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 61.69% (924) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_4382c | - | 0.0 | 64.49% (966) | putative membrane protein, MmpL |
| M. marinum M | MMAR_1005 | mmpL5 | 0.0 | 70.96% (954) | transmembrane transport protein, MmpL5 |
| M. avium 104 | MAV_2510 | - | 0.0 | 73.51% (955) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 69.81% (964) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_0757 | mmpL5 | 0.0 | 70.59% (959) | transmembrane transport protein, MmpL5 |
| M. vanbaalenii PYR-1 | Mvan_3189 | - | 0.0 | 73.24% (968) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0225|M.smegmatis_MC2_155 --MSTP-DAPTEAIPSAHG--KYTNHGG--IAKLIRTLAVPIILVWVALI
TH_3260|M.thermoresistible__bu -MTANPGHVRTDEAPTDRLDIKVSTHGPSFISTWIRRLSLPIIVGWIALI
Mflv_3435|M.gilvum_PYR-GCK -MSAPVSDARTDEFPAAHG-----PHRS-FIPRMIRLFAVPIILGWIALI
Mvan_3189|M.vanbaalenii_PYR-1 -MSAPVSDARTDEFPAARQ-----PHRP-FIPRMIRLFAVPIILGWIALI
Mb0695c|M.bovis_AF2122/97 -MIVQRTAAPTGSVPPDRH-----AARP-FIPRMIRTFAVPIILGWLVTI
Rv0676c|M.tuberculosis_H37Rv -MIVQRTAAPTGSVPPDRH-----AARP-FIPRMIRTFAVPIILGWLVTI
MMAR_1005|M.marinum_M -MTATTTDTPTDVVPPAEH-----PPRP-FIPRMIRTFAVPLILGWVLLI
MUL_0757|M.ulcerans_Agy99 -MTATTTDTPTDVVPPAEH-----PPRP-FIPRMIRTFAVPLILGWVLLI
MAV_2510|M.avium_104 -MSTTFDDTRTDAIPVVKEP----VREK--IPRFIRAFAVPIIIGWIALI
MAB_4382c|M.abscessus_ATCC_199 -MSKPMNDPTTDAFPAARH-----AARP-WLPRMIRVLAVPIVLGWIAVT
MLBr_02378|M.leprae_Br4923 MTVKCANDLDTHTKP------------P-FVARMIHRFAVPIILVWLAIA
* * :. *: :::*::: *:
MSMEG_0225|M.smegmatis_MC2_155 AVLNTIVPQLEEVGEMRSVSMSPDDAPSMIAMKRVGEVFEEFKSNSSVMI
TH_3260|M.thermoresistible__bu ALLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGELFDEGSSDSQVMI
Mflv_3435|M.gilvum_PYR-GCK VVVNVTVPQLETVGEAQAVSMSPNDAPSMISMKKVGELFEEGDSDSSVMI
Mvan_3189|M.vanbaalenii_PYR-1 VILNVTVPQLEAVGEARAVSMSPNEAPSLISMKKVGELFREGDSDSSVMI
Mb0695c|M.bovis_AF2122/97 AVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAMI
Rv0676c|M.tuberculosis_H37Rv AVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAMI
MMAR_1005|M.marinum_M AVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAMI
MUL_0757|M.ulcerans_Agy99 AVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAMI
MAV_2510|M.avium_104 AVLNVVVPQLDEVGKMRSVEMTPDDAQSVVATKRMGAVFNEYKSNSSVMI
MAB_4382c|M.abscessus_ATCC_199 IALNTVVPQLEEVGHMRAVSMSPNDAPAMIATKRVGEVFEEFKTSSSVMI
MLBr_02378|M.leprae_Br4923 VTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVFNEGNSDSVIMI
:. :*.*: **. ::*.::*. * : :: :::* :* * .:.* **
MSMEG_0225|M.smegmatis_MC2_155 VLEGEQPLGDEAHKYYDEIVDKLEADKTHIEHVQDFWGDPLTASGAQSSD
TH_3260|M.thermoresistible__bu VLEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWSDPLTATGVQSPD
Mflv_3435|M.gilvum_PYR-GCK VFEGDQPLGDEAHAWYDELVDRLEADTAHVQSVQDFWSDPLTASGSQSND
Mvan_3189|M.vanbaalenii_PYR-1 VFEGDQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDPLTASGSQSND
Mb0695c|M.bovis_AF2122/97 VLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSSD
Rv0676c|M.tuberculosis_H37Rv VLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSSD
MMAR_1005|M.marinum_M VLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSGD
MUL_0757|M.ulcerans_Agy99 VLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSGD
MAV_2510|M.avium_104 VLEGQQPLGADAHAYYDEIVRRLNADTKHVEHVQDMWSDPLTGAGAQSND
MAB_4382c|M.abscessus_ATCC_199 VLEGEQSLGADAHRFYDQMVRDLRADTTHVQHVQDFWADALTAAAAQSSD
MLBr_02378|M.leprae_Br4923 VLEGDKPLGDDAHRFYDGLIRKLRAD-KHVQSVQDFWGDPLTAPGAQSND
*:**:..** *: :** :: *.** *:: :**:*.*.**... ** *
MSMEG_0225|M.smegmatis_MC2_155 GLASYVQVYTAGNQGEALANESVKAVQDIVDSV---QAPPGVTAYVTGPA
TH_3260|M.thermoresistible__bu GKSAYVQVKLAGNQGEALANESVETVREMIATVSEEMAPEGVQAFVTGGA
Mflv_3435|M.gilvum_PYR-GCK GKAAYVQVKLSGNQGEALANESVKAVQQTVSSLE---PPPGVRAFVTGPA
Mvan_3189|M.vanbaalenii_PYR-1 GKAAYVQVKLAGNQGESLANESVQAAQEIVRSLE---PPPGVRAFVTGPA
Mb0695c|M.bovis_AF2122/97 GKAAYVQVKLAGNQGESLANESVEAVKTIVERLA---PPPGVKVYVTGSA
Rv0676c|M.tuberculosis_H37Rv GKAAYVQVKLAGNQGESLANESVEAVKTIVERLA---PPPGVKVYVTGSA
MMAR_1005|M.marinum_M GKAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---APPGVKVYVTGSA
MUL_0757|M.ulcerans_Agy99 GKAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---APPGVKVYVTGSA
MAV_2510|M.avium_104 GKASYVQVYLAGNQGEALANESVESVQNIVKSVQ---APNGVKAYVTGPA
MAB_4382c|M.abscessus_ATCC_199 GKAAYVQVYIAGDQGEALANESVEAVRHITAAVP---PSPGVRVYVTGPA
MLBr_02378|M.leprae_Br4923 GKAAYVQLSLAGNQGELLAQESIDAVRKIVVQTP---APPGITAWVTGAS
* ::***: :*:*** **:**:.:.: .. *: .:*** :
MSMEG_0225|M.smegmatis_MC2_155 AMSADQQIAGDRSMRMIEALTFTVIIIMLLMVYRSIITVILSLVMVVFSL
TH_3260|M.thermoresistible__bu AMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVITVLIALFMVLIGL
Mflv_3435|M.gilvum_PYR-GCK ALAADQHIAGDRSMRLIEAATFTVIIVMLLLVYRSIVTVLITLVMVVMSL
Mvan_3189|M.vanbaalenii_PYR-1 ALAADQHIASDRSVRVIELVTFAVIIVMLLLVYRSIVTVLLTLVMVVLSL
Mb0695c|M.bovis_AF2122/97 ALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTMVVLGL
Rv0676c|M.tuberculosis_H37Rv ALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTMVVLGL
MMAR_1005|M.marinum_M ALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTMVVLGL
MUL_0757|M.ulcerans_Agy99 ALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTMVVLGL
MAV_2510|M.avium_104 ALSADQHTAGDRSLQLITAATFTVIIGMLLLVYRSVITVLLTLVMVVLEL
MAB_4382c|M.abscessus_ATCC_199 TTSADQEIVANDSMELIELVTFGVIMVMLLLVYRSVITMVIVVAMVMLEL
MLBr_02378|M.leprae_Br4923 ALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITVILLLFTVGIES
: ** . .: *: * :. ** **:****.:* : : * :
MSMEG_0225|M.smegmatis_MC2_155 AAARGVIAFLGYYEIIGLSTFATNLLVTLAIAAATDYAIFLIGRYQEARS
TH_3260|M.thermoresistible__bu LTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYAIFLIGRYQEARS
Mflv_3435|M.gilvum_PYR-GCK ATARGVVAFLGHHEIIGLSTFATNLLVTLAIAAATDYAIFLIGRYQEART
Mvan_3189|M.vanbaalenii_PYR-1 ATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIFLIGRYQEART
Mb0695c|M.bovis_AF2122/97 LATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEARG
Rv0676c|M.tuberculosis_H37Rv LATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEARG
MMAR_1005|M.marinum_M LATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEARG
MUL_0757|M.ulcerans_Agy99 LATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEARG
MAV_2510|M.avium_104 SAARGMVAFLGYYKIIGLSTFATNLLVTLAIAAATDYAIFLIGRYQEARA
MAB_4382c|M.abscessus_ATCC_199 ASARGLVALLSYHNAFGLTSFATSLVVTLAIAAATDYAIFLIGRYQEARR
MLBr_02378|M.leprae_Br4923 AVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGIFITGRYQEARQ
:** :*:*. :**: **..*:. **** ..**.**: *******
MSMEG_0225|M.smegmatis_MC2_155 VGESREQAYYTMFHSTAHVVLGSGMTIAGATLCLHFTRMPYFQSLGIPLA
TH_3260|M.thermoresistible__bu VGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTRLPYFQSLGPPLA
Mflv_3435|M.gilvum_PYR-GCK AGEDKETAYYTMFHGTAHVVLGSGLTIAGATFCLSFTRLPYFQTMGVPLA
Mvan_3189|M.vanbaalenii_PYR-1 NGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLPYFQTLGVPLA
Mb0695c|M.bovis_AF2122/97 LGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPLA
Rv0676c|M.tuberculosis_H37Rv LGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPLA
MMAR_1005|M.marinum_M LGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPLA
MUL_0757|M.ulcerans_Agy99 LGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPLA
MAV_2510|M.avium_104 VGESREDAYYTMYKGTAHVVAGSGMTIAGATFCLHFTNLPYFQTLGIPLA
MAB_4382c|M.abscessus_ATCC_199 AGEDRESAYYTMFHGTAHVVLASGLTIAGATFCLHFTRLPYFKTTGIPLS
MLBr_02378|M.leprae_Br4923 ANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARMPYFQTLGVPCA
. .: *::**: .* **: .**:**:*** ** *:.:***:: * * :
MSMEG_0225|M.smegmatis_MC2_155 IGMTVVVLTALAVGPAIITVASRFGKTLEPKRAMRTRGWRKVGAAVVRWP
TH_3260|M.thermoresistible__bu IGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARVRFWRKVGASVVRWP
Mflv_3435|M.gilvum_PYR-GCK IGMFVVVMAGVVLMVACISVATRFGKLLEPKRAMRIRGWRKIGAAIVRWP
Mvan_3189|M.vanbaalenii_PYR-1 IGMFVVVMAGVILMVAMISVATRFGKLLEPKRAMRIRGWRKIGAAVVRWP
Mb0695c|M.bovis_AF2122/97 IGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAAIVRWP
Rv0676c|M.tuberculosis_H37Rv IGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAAIVRWP
MMAR_1005|M.marinum_M IGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAAIVRWP
MUL_0757|M.ulcerans_Agy99 IGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAAIVRWP
MAV_2510|M.avium_104 IGMVVVVAAALTLGPAVISVASRFRQTLEPRRTQRIRGWRKVGTVVVRWP
MAB_4382c|M.abscessus_ATCC_199 IGMAIVVAMALTLGPALISVASRFGNVLEPKGAGKARGWRRVGAATVRWP
MLBr_02378|M.leprae_Br4923 VGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKVRGWRRIGTVVVRWP
:** : : .: : * ::* :** :**: : * **::*: ****
MSMEG_0225|M.smegmatis_MC2_155 GPILVAAVGLSMIGLLALPGYRTNYNDRDYQPADLPANSGYAAADRHFSQ
TH_3260|M.thermoresistible__bu GPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSNAGYEAAERHFDP
Mflv_3435|M.gilvum_PYR-GCK GPILVATMAITLVGLLALPGYKTNYNDRDYLPADLPANQGYAVADEHFDQ
Mvan_3189|M.vanbaalenii_PYR-1 GPILVATLAITLVGLLALPGYKTNYNDRTYLPADLPANEGYAVADRHFDQ
Mb0695c|M.bovis_AF2122/97 GPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAERHFSQ
Rv0676c|M.tuberculosis_H37Rv GPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAERHFSQ
MMAR_1005|M.marinum_M GPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAERHFSQ
MUL_0757|M.ulcerans_Agy99 GPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAERHFSQ
MAV_2510|M.avium_104 GPILVLTIGVALVGLLTLPGYRTNYNDRNYLPTDLPANEGYAAADRHFSQ
MAB_4382c|M.abscessus_ATCC_199 GAILVAAVTMALVGLLTLPGYHTTYNDRIYLPGQVPANVGYAAADRHFSK
MLBr_02378|M.leprae_Br4923 LPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQGFAAADRHFPQ
.*:: : ::::***:****:*.*:** * * ..*:* *: .*:.**
MSMEG_0225|M.smegmatis_MC2_155 ARMNPELLMIESDHDLRNSADFLVIERIAKRVVGVPGVSRVQAITRPQGT
TH_3260|M.thermoresistible__bu ARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGISRVQGITRPEGT
Mflv_3435|M.gilvum_PYR-GCK ARMNPELLMIESDHDLRNSADFLVIDKIAKALFRVEGIARVQAITRPDGK
Mvan_3189|M.vanbaalenii_PYR-1 ARMNPELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIARVQAITRPDGK
Mb0695c|M.bovis_AF2122/97 ARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAITRPDGK
Rv0676c|M.tuberculosis_H37Rv ARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAITRPDGK
MMAR_1005|M.marinum_M ARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAITRPDGK
MUL_0757|M.ulcerans_Agy99 ARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAITRPDGK
MAV_2510|M.avium_104 ARMNPEVLMIESDHDLRNSADFLVIDKIAKTVFRVPGIGRVQAITRPQGT
MAB_4382c|M.abscessus_ATCC_199 AKMNPDLLLVESDHDLRNPADFLVIDKIAKALARVRGIAQVQTITRPDGK
MLBr_02378|M.leprae_Br4923 ARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGISRVQAITRPDGT
*:::*::*::*****:**.:***:::::*: : * *:.:** ****:*.
MSMEG_0225|M.smegmatis_MC2_155 PIEHTSIPFTISMQGTTQTMNQKYMQDRMADMLVQADQMQVTVDTMTKMI
TH_3260|M.thermoresistible__bu PIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADEMQLMIDTMDKMI
Mflv_3435|M.gilvum_PYR-GCK PIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIKTMEKMS
Mvan_3189|M.vanbaalenii_PYR-1 PIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIATMEKMS
Mb0695c|M.bovis_AF2122/97 PIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQMERMH
Rv0676c|M.tuberculosis_H37Rv PIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQMERMH
MMAR_1005|M.marinum_M PIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQMERMH
MUL_0757|M.ulcerans_Agy99 PIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQMERMH
MAV_2510|M.avium_104 PIEHTSIPFQISMQGVTQQMNQKYQQDQMADMLHQADMMQTTIDSMEKMQ
MAB_4382c|M.abscessus_ATCC_199 PIKHTSIPFNMSTANTAQTLTNDYSQANFANLLKQADAMQVTIDSMEKMQ
MLBr_02378|M.leprae_Br4923 AMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENMAETIASMRRMH
.:.*:**** :* .. * . .* : :* *.: : : * :*
MSMEG_0225|M.smegmatis_MC2_155 ALMEQMRDTMNSMVGKMHGMVDDIKELRDNISDFDDFFRPIRNYLYWEPH
TH_3260|M.thermoresistible__bu ALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFFRPIRNYFYWEPH
Mflv_3435|M.gilvum_PYR-GCK GLTAQMADITHSMVTKMEGMVVDIEDLRDSIANFDDFFRPIRNYFYWEPH
Mvan_3189|M.vanbaalenii_PYR-1 NLTAQMADITHSMVSKMENMLTDIEDLRDSIANFDDFFRPIRNYFYWEPH
Mb0695c|M.bovis_AF2122/97 SLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYFYWEKH
Rv0676c|M.tuberculosis_H37Rv SLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYFYWEKH
MMAR_1005|M.marinum_M DLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYFYWEKH
MUL_0757|M.ulcerans_Agy99 DLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYFYWEKH
MAV_2510|M.avium_104 SITVQMSNDMHVMVQKMHDMTIDINDLRNKMADFEDFFRPMRSYFYWEKH
MAB_4382c|M.abscessus_ATCC_199 AITQQMAEVTASMSANMKNTAQDINQLRDNMSNFDDQFRPIRNYFYWEKH
MLBr_02378|M.leprae_Br4923 QLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWRPIRSYFYWERH
: : : : . .:*: :::*:* :**:*.*:*** *
MSMEG_0225|M.smegmatis_MC2_155 CYNIPMCYSLRSIFDTMDGIDTMTADFEQIVPDMDRMNALIPVMIANMEP
TH_3260|M.thermoresistible__bu CYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLNDLMPQMLTLLPT
Mflv_3435|M.gilvum_PYR-GCK CYNIPVCWALRSVFDTLDGINIMTDDFKAIIPDMKRLDTLMPQMVALMPE
Mvan_3189|M.vanbaalenii_PYR-1 CYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDSLMPQMVALMPE
Mb0695c|M.bovis_AF2122/97 CYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLTAMMPE
Rv0676c|M.tuberculosis_H37Rv CYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLTAMMPE
MMAR_1005|M.marinum_M CFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMTAMMPE
MUL_0757|M.ulcerans_Agy99 CFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMTAMMPE
MAV_2510|M.avium_104 CYDIPVCWSLRSVFDALDGIDTMTDDIQSLLPIMDHLDTLMPQMVALMPS
MAB_4382c|M.abscessus_ATCC_199 CFDIPMCWALRSVFDALDGISLMSDDFDTLVPDIQRMAGLMPQMVVLMPP
MLBr_02378|M.leprae_Br4923 CFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDLLMPQMLEQFPP
*::**:*:::**:**::**:. : : :: :..: *:* : :
MSMEG_0225|M.smegmatis_MC2_155 MITTMKTMKTMMLTMQATNAGLQDQMAAMQENSTAMGEAFDKAKNDDSFY
TH_3260|M.thermoresistible__bu TVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGEAFDQARNDDSFY
Mflv_3435|M.gilvum_PYR-GCK MIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMNDDSFY
Mvan_3189|M.vanbaalenii_PYR-1 MIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMNDDSFY
Mb0695c|M.bovis_AF2122/97 MIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRNDDSFY
Rv0676c|M.tuberculosis_H37Rv MIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRNDDSFY
MMAR_1005|M.marinum_M MIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRNDDSFY
MUL_0757|M.ulcerans_Agy99 MIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRNDDSFY
MAV_2510|M.avium_104 MIENMKAMKTTMLTMYSTQKGLQDQQNEAQKNSNAMGKAFDASKNDDSFY
MAB_4382c|M.abscessus_ATCC_199 MIASMKEQKQIMLSQYQVQKAQQDQTMAMQANASAMGQAFDDAKNDDSFY
MLBr_02378|M.leprae_Br4923 MIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKAFDTAKNDDSFY
: .*: : :*: . ** . :: :**:*** : ******
MSMEG_0225|M.smegmatis_MC2_155 LPPETFDNPDFKRGMKMFLSPDGHAVRFIISHEGDPMSPEGIQKIDDIKL
TH_3260|M.thermoresistible__bu LPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMTPEGLAKIEPIKK
Mflv_3435|M.gilvum_PYR-GCK LPPEIFENADFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKIDKIKT
Mvan_3189|M.vanbaalenii_PYR-1 LPPEIFENKDFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKIDKIKT
Mb0695c|M.bovis_AF2122/97 LPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARIAKIKT
Rv0676c|M.tuberculosis_H37Rv LPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARIAKIKT
MMAR_1005|M.marinum_M LPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHIDKIKT
MUL_0757|M.ulcerans_Agy99 LPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHIDKIKT
MAV_2510|M.avium_104 LPPETFNNKEFKKGMKNFISPDGHAVRFIISHDGDPMTQEGISHIGAIKK
MAB_4382c|M.abscessus_ATCC_199 LPPEAFETADFKSGIKLFVSPNGHAVRFTIFHQSDPLTEEGTSRVEPLKT
MLBr_02378|M.leprae_Br4923 LPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASVAGIASINAIRT
**** *.. :*: .:: *:*.:***.** * * .** : * : ::
MSMEG_0225|M.smegmatis_MC2_155 AAKEAIKGTPLEGSKIYLGGTAATFKDMQEGANYDLIIAGIAALCLIFII
TH_3260|M.thermoresistible__bu AAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLIIAAIAAVCLIFII
Mflv_3435|M.gilvum_PYR-GCK AAKEAVKGTPLEGSKIYLGGTAAVFKDMQDGNNYDLLIAGIAALGLIFII
Mvan_3189|M.vanbaalenii_PYR-1 AAKEAIKGTPLEGSKIYLGGTAATFKDMQDGNNYDLLIAGISALGLIFII
Mb0695c|M.bovis_AF2122/97 AAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALCLIFII
Rv0676c|M.tuberculosis_H37Rv AAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALCLIFII
MMAR_1005|M.marinum_M AAKEAIKGTPLEGSTIYLGGTAAMFKDLSDGNTFDLMIAGISALCLIFII
MUL_0757|M.ulcerans_Agy99 AAKEAIKGTPLEGSTIYLGGTVAMFKDLSDGNTFDLMIAGISALCLIFII
MAV_2510|M.avium_104 AAYEALKGTPLEGSKIYLAGTASIYKDLSDGNNYDLLIAGISSLCLIFII
MAB_4382c|M.abscessus_ATCC_199 AAEDAIKGTPLEGSSIYVGGSAALYKDMQEGADYDLLIAAVAALILIFLI
MLBr_02378|M.leprae_Br4923 AAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIAGISSLCLIFII
** :*:******.: :*:.*:.: :**:.:* :**:**.:::: ***:*
MSMEG_0225|M.smegmatis_MC2_155 MLIITRAVVASAVIVGTVVISLGSAFGLSVLIWQHLVGLELHWMVLAMAV
TH_3260|M.thermoresistible__bu MLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVGLEMHWMVFPMAL
Mflv_3435|M.gilvum_PYR-GCK MLILTRAVVAAAVIVGTVVLSLAASFGLSVLFWQHILGQPLHWMVLAMAV
Mvan_3189|M.vanbaalenii_PYR-1 MLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGTELHWMVLAMAV
Mb0695c|M.bovis_AF2122/97 MLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMAV
Rv0676c|M.tuberculosis_H37Rv MLITTRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMAV
MMAR_1005|M.marinum_M MLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMAV
MUL_0757|M.ulcerans_Agy99 MLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHVLGIELHWLVLAMAV
MAV_2510|M.avium_104 MLIITRGVVASAVIVGTVLLSLGASFGLSVLIWQHLIGIELHWMVLAMSV
MAB_4382c|M.abscessus_ATCC_199 MVLLTRAVVAAAVIVGTVVLSLGASFGLSVLLWQHIIGTPLHWMVLPMAV
MLBr_02378|M.leprae_Br4923 MLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGIELHYLVLAMSV
*:: **..**:******* :**.::**:***:**:::* :*::*:.*::
MSMEG_0225|M.smegmatis_MC2_155 IVLLAVGADYNLLLVSRIKEEIHAGLDTGMIRAMGGSGSVVTAAGLVFAF
TH_3260|M.thermoresistible__bu IILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSGSVVTAAGLVFGL
Mflv_3435|M.gilvum_PYR-GCK IILLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAGLVFAL
Mvan_3189|M.vanbaalenii_PYR-1 IVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAGLVFAL
Mb0695c|M.bovis_AF2122/97 IILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFAF
Rv0676c|M.tuberculosis_H37Rv IILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFAF
MMAR_1005|M.marinum_M IILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFAF
MUL_0757|M.ulcerans_Agy99 IILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFAF
MAV_2510|M.avium_104 IILLAVGADYNLLLVARFKEEIHAGLNTGIIRSMGGTGSVVTSAGLVFAF
MAB_4382c|M.abscessus_ATCC_199 IILLAVGADYNLLLVSRMKEELHAGLHTGTIRSMAGTGSVVTSAGLVFAF
MLBr_02378|M.leprae_Br4923 IVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGKVVTNAGLVFAF
*:*****:*****:*:*:::*: **: ** **:*.*:*.*** *****.:
MSMEG_0225|M.smegmatis_MC2_155 TMMSMAVSELAIIGQVGTTIGLGLLFDTLVIRSFMTPSIAALLGKWFWWP
TH_3260|M.thermoresistible__bu TMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPSIATLLGRWFWWP
Mflv_3435|M.gilvum_PYR-GCK TMMSMSVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGPWFWWP
Mvan_3189|M.vanbaalenii_PYR-1 TMMSMAVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGPWFWWP
Mb0695c|M.bovis_AF2122/97 TMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWWP
Rv0676c|M.tuberculosis_H37Rv TMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWWP
MMAR_1005|M.marinum_M TMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWWP
MUL_0757|M.ulcerans_Agy99 TMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWWP
MAV_2510|M.avium_104 TMMTMAVSELTVIGQVGTTIGLGLLFDTLIVRSLMTPSIAALLGKWFWWP
MAB_4382c|M.abscessus_ATCC_199 TMMGMMVSSFTVIGQIGTTIGLGLLFDTLVVRSFMTPSIASLLGRWFWWP
MLBr_02378|M.leprae_Br4923 TMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSIAALLGRWFWWP
** : **.: ::.*:*****:******:::*::* ****:*** *****
MSMEG_0225|M.smegmatis_MC2_155 QMVRRRPKPSPWPKPIQREPADAVN---
TH_3260|M.thermoresistible__bu QFVRKRPVPQPWPSVPPRGPATTDRVSV
Mflv_3435|M.gilvum_PYR-GCK QRVRTRPVPAPWPRSGGLQSDPSEGVRA
Mvan_3189|M.vanbaalenii_PYR-1 QRVRTRPVPAPWPRPGGLQSDPSEGVKV
Mb0695c|M.bovis_AF2122/97 QVVRQRPVPQPWPSPASARTFALV----
Rv0676c|M.tuberculosis_H37Rv QVVRQRPIPQPWPSPASARTFALV----
MMAR_1005|M.marinum_M QVVRQRPAPQPWPTPAPTP--ATTSV--
MUL_0757|M.ulcerans_Agy99 QVVRQRPAPQPWPTPAPTPPPATTSV--
MAV_2510|M.avium_104 QRVRQRPVPSPWPKPAAQEREVLADA--
MAB_4382c|M.abscessus_ATCC_199 MVVRPRPRPQPWPQPLHAGAAEAVRA--
MLBr_02378|M.leprae_Br4923 QQGRTRPLLTVAAPVGLPPDRSLVYSD-
* ** .