For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRVLITGADTKLGVAVNDHFTARGAVTAAPPADTDHADRAAVRDAVRRAVERMGGIDVLVVAHGLPHVAA FTEQSMAEFWKHVDTTLTGSFLYAQAAAEHMRDNGTGGRIVLTTSRWHVGGAKLSAVATAAGGIVALTKT LTRDFGAFGIGVNAVAIGNVDSEWSVCDLEGATPPLDRIGSVDQVAGVIGLLSRPELKAAVGQIVNADGG QSRNRV*
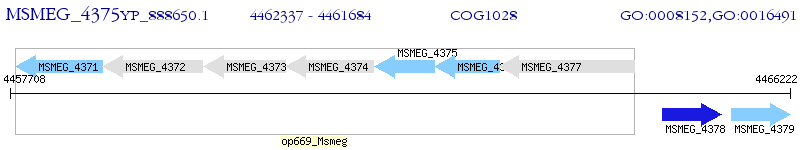
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4375 | - | - | 100% (217) | 3-oxoacyl- |
| M. smegmatis MC2 155 | MSMEG_1071 | - | 4e-24 | 31.56% (244) | 3-oxoacyl- |
| M. smegmatis MC2 155 | MSMEG_6538 | - | 3e-19 | 29.82% (228) | 3-oxoacyl-(acyl-carrier-protein) reductase, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1519 | fabG1 | 3e-16 | 27.75% (227) | 3-oxoacyl- |
| M. gilvum PYR-GCK | Mflv_0798 | - | 2e-24 | 32.79% (244) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1483 | fabG1 | 3e-16 | 27.75% (227) | 3-oxoacyl- |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-16 | 26.67% (225) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_2723c | - | 2e-17 | 28.89% (225) | 3-oxoacyl- |
| M. marinum M | MMAR_2289 | fabG1 | 5e-17 | 28.19% (227) | 3-oxoacyl- |
| M. avium 104 | MAV_1395 | - | 2e-18 | 27.15% (221) | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2435 | - | 8e-17 | 27.93% (222) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1491 | fabG1 | 1e-16 | 27.75% (227) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_0045 | - | 1e-26 | 34.84% (244) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2289|M.marinum_M MTDTATEQNTESAADGGKPAFVSRSVLVTGGNRGIGLAIAQRLAADGHRV
MUL_1491|M.ulcerans_Agy99 MTDTATEQNTESAADGGKPAFVSRSVLVTGGNRGIGLAIAQRLATDGHRV
Mb1519|M.bovis_AF2122/97 MTATATEG--------AKPPFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
Rv1483|M.tuberculosis_H37Rv MTATATEG--------AKPPFVSRSVLVTGGNRGIGLAIAQRLAADGHKV
MLBr_01807|M.leprae_Br4923 MTDAAAKVS--AAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRV
MAB_2723c|M.abscessus_ATCC_199 MTNA---------------EFVPRSVLVTGGNRGIGLAVAQRLAADGHKV
MAV_1395|M.avium_104 ------------------------MAVVTGGGSGIGRAIVERLAHDGHRV
TH_2435|M.thermoresistible__bu ---------------------------VTGGASGLGRAIAARLGDDGHRV
Mflv_0798|M.gilvum_PYR-GCK ----------------MTTDLRGRVALVTGAAQGMGAAHARRLAKAGATV
Mvan_0045|M.vanbaalenii_PYR-1 ----------------MSTDLRGRVALVTGAARGMGAAHARRLAAAGATV
MSMEG_4375|M.smegmatis_MC2_155 ----------------------MTRVLITGADTKLGVAVNDHFTARGAVT
:**. :* * :: * .
MMAR_2289|M.marinum_M AVTHRGSGAPEGLFG-----------VECDVTDNDAVDRAFKEVEEHQGP
MUL_1491|M.ulcerans_Agy99 AVTHRGSGAPEGLFG-----------VECDVTDNDAVDRAFKEVEEHQGP
Mb1519|M.bovis_AF2122/97 AVTHRGSGAPKGLFG-----------VECDVTDSDAVDRAFTAVEEHQGP
Rv1483|M.tuberculosis_H37Rv AVTHRGSGAPKGLFG-----------VECDVTDSDAVDRAFTAVEEHQGP
MLBr_01807|M.leprae_Br4923 AVTHRASVVPDWFFG-----------VECDVTDNDAIGAAFKAVEEHQGP
MAB_2723c|M.abscessus_ATCC_199 AVTHRGSGAPEGLLG-----------VQCDITDNDQIDAAFKEVEEKQGP
MAV_1395|M.avium_104 AVLDVNEEAAEKVAARVAADGAHAIAVPTDVAESASVAAAFESVRRALGS
TH_2435|M.thermoresistible__bu AVLDIDHAAAEQTAAELRAHGGDAVAVGADVADQDAVHDAFAAIRGSLGP
Mflv_0798|M.gilvum_PYR-GCK AVNDREDSDALAALASEIG----GLTAAGDVSDPAQCRAVTARIAETAGR
Mvan_0045|M.vanbaalenii_PYR-1 AVNDREDSDALTALATEIG----GLTAPGDVSDPKSCRALATHVAATTGR
MSMEG_4375|M.smegmatis_MC2_155 AAPP----------------------ADTDHADRAAVRDAVRRAVERMGG
*. . * :: *
MMAR_2289|M.marinum_M VEVLVSNAGLSADAFLIRMTEERFEKVIDANLTGAFRVAQRASRSMQRKK
MUL_1491|M.ulcerans_Agy99 VEVLVSNAGLSADAFLIRMTEERFEKVIDANLTGAFRVAQRASRSMQRKK
Mb1519|M.bovis_AF2122/97 VEVLVSNAGLSADAFLMRMTEEKFEKVINANLTGAFRVAQRASRSMQRNK
Rv1483|M.tuberculosis_H37Rv VEVLVSNAGLSADAFLMRMTEEKFEKVINANLTGAFRVAQRASRSMQRNK
MLBr_01807|M.leprae_Br4923 VEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
MAB_2723c|M.abscessus_ATCC_199 VEVLVANAGITADMLIMRMSEEQFLKVIDANLIGTFRLAKRASRNMIKKR
MAV_1395|M.avium_104 VQVLVTSAAITGFKPFAEITIEDWNRHLAVNLTGTFLCLQAALPDMVEAG
TH_2435|M.thermoresistible__bu VHILVTSAAIAGFTRFDEITLDEWNRYLAVNLTGTFLCVQAALTDMVADG
Mflv_0798|M.gilvum_PYR-GCK LDILVANHAYMTMAPLLEHDPDDWWKVVDTNLGGTFFLVQSVLPYMREAG
Mvan_0045|M.vanbaalenii_PYR-1 LDVLVANHAYMTMAPLLEHADDDWWKVVDTNLGGTFFLVQSVLPHMRRTG
MSMEG_4375|M.smegmatis_MC2_155 IDVLVVAHGLPHVAAFTEQSMAEFWKHVDTTLTGSFLYAQAAAEHMRDNG
:.:** . : . : : : ..* * * : . *
MMAR_2289|M.marinum_M FGRLIFIGSVSGSWGIGNQANYAASKAGVIGMARSIARELSKVNVTANVV
MUL_1491|M.ulcerans_Agy99 FGRLIFIGSVSGSWGIGNQANYAASKAGVIGMARSIARELSKVNVTANVV
Mb1519|M.bovis_AF2122/97 FGRMIFIGSVSGSWGIGNQANYAASKAGVIGMARSIARELSKANVTANVV
Rv1483|M.tuberculosis_H37Rv FGRMIFIGSVSGSWGIGNQANYAASKAGVIGMARSIARELSKANVTANVV
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVV
MAB_2723c|M.abscessus_ATCC_199 FGRLIFMGSVAGQIGLPGQANYSASKAGLVGMARALTRELGSRSITANVV
MAV_1395|M.avium_104 WGRVVTISSTAAQTGSPRQGHYSASKGGVIALTRTIALEYAAHGITANTV
TH_2435|M.thermoresistible__bu WGRIVTISSAAGQQGAHRQGHYAATKGGVIALTKTVALDYAAKGITANTV
Mflv_0798|M.gilvum_PYR-GCK AGRIVVISSEWGVTGWPGATAYAASKSGLISLVKTLGRELAREHIIVNAV
Mvan_0045|M.vanbaalenii_PYR-1 AGRIVVIASEWGVTGWPEATAYAAAKSGLISLVKTLGRELAREHIIVNAV
MSMEG_4375|M.smegmatis_MC2_155 TGGRIVLTTSRWHVGGAKLSAVATAAGGIVALTKTLTRDFGAFGIGVNAV
* : : : * ::: .*::.:.::: : . : .*.*
MMAR_2289|M.marinum_M APGYIDTDMTRALDER-------IQEGALQFIPAKRVGTAAEVAGVVSFL
MUL_1491|M.ulcerans_Agy99 APGYIDTDMTRALDER-------IQEGALQFIPAKRVGTAAEVAGVVSFL
Mb1519|M.bovis_AF2122/97 APGYIDTDMTRALDER-------IQQGALQFIPAKRVGTPAEVAGVVSFL
Rv1483|M.tuberculosis_H37Rv APGYIDTDMTRALDER-------IQQGALQFIPAKRVGTPAEVAGVVSFL
MLBr_01807|M.leprae_Br4923 APGFIDTEMTRAMDSK-------YQEAALQAIPLGRIGAVEEIAGAVSFL
MAB_2723c|M.abscessus_ATCC_199 APGFIDTDMTRELGEK-------HVETALTAIPMGRIGEPSEVAGVVSWL
MAV_1395|M.avium_104 PPFSVDTPMLRAAQEAG---NLPPVKYLAKASPVGRLGTGEDIAAACAFL
TH_2435|M.thermoresistible__bu PPFVIDSPMLREQQRRG---TLPPAEYLTRAIPMGRLGVAEDVAAVCSFL
Mflv_0798|M.gilvum_PYR-GCK APGVIDTPQLQVDADAAGVALEVIHRRYAESIPLGRIGVGDEVSAAVELL
Mvan_0045|M.vanbaalenii_PYR-1 APGVIDTPQLQVDADAAGVGLDDIHRRYAEAIPLGRIGSADEVAVAVELL
MSMEG_4375|M.smegmatis_MC2_155 AIGNVDSEWSVCDLEG-------------ATPPLDRIGSVDQVAGVIGLL
. :*: * *:* ::: . *
MMAR_2289|M.marinum_M ASEDASYISGAVIPVDGGMGMGH-
MUL_1491|M.ulcerans_Agy99 ASEDASYISGAVIPVDGGMGMGH-
Mb1519|M.bovis_AF2122/97 ASEDASYISGAVIPVDGGMGMGH-
Rv1483|M.tuberculosis_H37Rv ASEDASYISGAVIPVDGGMGMGH-
MLBr_01807|M.leprae_Br4923 ASEDAGYISGAVIPVDGGLGMGH-
MAB_2723c|M.abscessus_ATCC_199 ASPDAAYVSGAVIPVDGGLGMGH-
MAV_1395|M.avium_104 CSDEAGYITGQIIGVNGGEVI---
TH_2435|M.thermoresistible__bu CSDAAGYLTGQVIGVNGGAIR---
Mflv_0798|M.gilvum_PYR-GCK ADFDVEAIVGQVISCNGGSMRARA
Mvan_0045|M.vanbaalenii_PYR-1 SDFDLEAVVGQVISCNGGSTRTRA
MSMEG_4375|M.smegmatis_MC2_155 SRPELKAAVGQIVNADGGQSRNRV
. * :: :**