For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKLEGKSAVVAAGAKNLGGLISTTLASRGVNVAVHYNSASTEADADKTVAAVQDAGVKAVKVQGDLTVPA NVEKLFDTAIDAFGGVDIAINTVGKVLRKPIAETTEAEYDSMFDINSKAAYFFLQQAGKRLNDNGSITTI VTSLLAAYTDGYSTYAGSKSPVEHFTRAASKEFGVRGINVNNIAPGPMDTPFFYPEETPERVEFHKSQAM GNRLTDIGDIAPLAVFLADEGHWINGQTLFTNGGYTTR*
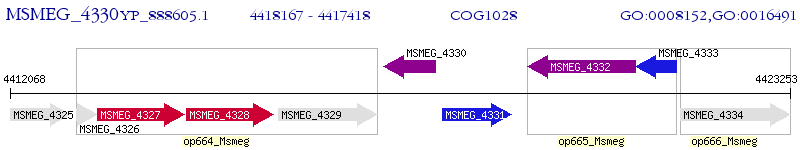
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4330 | - | - | 100% (249) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5300 | - | 6e-34 | 33.33% (237) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2029 | - | 3e-29 | 33.87% (248) | 3-ketoacyl-ACP/CoA redutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 6e-22 | 28.97% (252) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1235 | - | 2e-24 | 30.24% (248) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0927c | - | 6e-22 | 28.97% (252) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 4e-12 | 29.23% (195) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4657c | - | 2e-77 | 57.14% (252) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4033 | fabG | 2e-22 | 30.12% (249) | 3-oxoacyl- |
| M. avium 104 | MAV_1572 | - | 3e-21 | 28.11% (249) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3122 | - | 1e-112 | 79.92% (239) | PUTATIVE short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_4442 | - | 7e-21 | 28.40% (250) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5843 | - | 1e-25 | 32.52% (246) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4033|M.marinum_M --------------------------------------------------
MAV_1572|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mflv_1235|M.gilvum_PYR-GCK --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4330|M.smegmatis_MC2_155 --------------------------------------------------
TH_3122|M.thermoresistible__bu --------------------------------------------------
MAB_4657c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5843|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4033|M.marinum_M --------------------------------------------------
MAV_1572|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mflv_1235|M.gilvum_PYR-GCK --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4330|M.smegmatis_MC2_155 --------------------------------------------------
TH_3122|M.thermoresistible__bu --------------------------------------------------
MAB_4657c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5843|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4033|M.marinum_M -------------------------------------------MSVSL--
MAV_1572|M.avium_104 ---------------------------------------------MSL--
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mflv_1235|M.gilvum_PYR-GCK --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4330|M.smegmatis_MC2_155 --------------------------------------------------
TH_3122|M.thermoresistible__bu --------------------------------------------------
MAB_4657c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5843|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4033|M.marinum_M --------------------------------------------------
MAV_1572|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mflv_1235|M.gilvum_PYR-GCK --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4330|M.smegmatis_MC2_155 --------------------------------------------------
TH_3122|M.thermoresistible__bu --------------------------------------------------
MAB_4657c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5843|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4033|M.marinum_M ----------LNGQIAVVTGGAQGLGLAIAERFVSEGAR---VVLGDVN-
MAV_1572|M.avium_104 ----------LSGQTAVITGGAQGLGFAIAERFVAEGAR---VVLGDVN-
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGAR---VVAIDVES
Mflv_1235|M.gilvum_PYR-GCK ------MFTSLQGRSAAVTGGSKGIGRGIAETFARAGIN---VVITART-
Mb0950c|M.bovis_AF2122/97 ---MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGAD---VLIASRT-
Rv0927c|M.tuberculosis_H37Rv ---MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGAD---VLIASRT-
MUL_4442|M.ulcerans_Agy99 ---MILDRFRLDDQVAVITGGGRGFGAAIALAFAEAGAD---VVIASRT-
MSMEG_4330|M.smegmatis_MC2_155 -------MSKLEGKSAVVAAGAKNLGGLISTTLASRGVN-VAVHYNSAST
TH_3122|M.thermoresistible__bu -----------------VAAGAKNLGGLISTTLAERGVN-VAVHYHSAAT
MAB_4657c|M.abscessus_ATCC_199 -----MTDHSLNGKTVLITGGGKNLGGLIARDLAANGAGAIAIHYNSPSS
Mvan_5843|M.vanbaalenii_PYR-1 -----MSTAPLAGKRALVTGGSRGIGAEIVRRLAADGAA---VAFTYAAS
::...:.:* * .. * :
MMAR_4033|M.marinum_M -LEATEAAAKQLGGGEVAVAVRCDVTKADEVETLLQTALERFGG-LDIMV
MAV_1572|M.avium_104 -LEATQTAAKQLGGDQVALAVRCDVTKSSEVETLIQTAVERFGG-LDIMV
MLBr_02565|M.leprae_Br4923 AAEALDETANRVGG----TTLSLDVTADDAVDKITEHLHDYQGGHADILV
Mflv_1235|M.gilvum_PYR-GCK QDDIDATVADLAGLAGTVTGVAADVTSPEDCRRVVETAIERHGG-LDIVC
Mb0950c|M.bovis_AF2122/97 SSELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFGK-LDIVV
Rv0927c|M.tuberculosis_H37Rv SSELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFGK-LDIVV
MUL_4442|M.ulcerans_Agy99 QSQLDEVADTVRSIGRRAHVVAADLAHPEATAELAGQAVEAFGR-LDIVV
MSMEG_4330|M.smegmatis_MC2_155 EADADKTVAAVQDAGVKAVKVQGDLTVPANVEKLFDTAIDAFGG-VDIAI
TH_3122|M.thermoresistible__bu EPDADKTVAAVEATGARAIKVQGDLTVPANVTRLFDATVEAFGG-VDIAV
MAB_4657c|M.abscessus_ATCC_199 KDAAEETVAAVQAAGAKAVAFQGDLTTGDAVSKLFADTVAAVGR-PDIAI
Mvan_5843|M.vanbaalenii_PYR-1 AAHAEKVVAEVAASGGEAVAIRADSADPTQVADAVEQTVAELGG-LDILV
. . * : * **
MMAR_4033|M.marinum_M NNAGITRDATMRKMTEEQFDQVIDVHLKGTWNGIRLAAAIMREN-KRGAI
MAV_1572|M.avium_104 NNAGITRDATMRKMTEEQFDQVIAVHLKGTWNGTRLAAAIMREN-KRGAI
MLBr_02565|M.leprae_Br4923 NNAGITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSIS-KGGRV
Mflv_1235|M.gilvum_PYR-GCK PNAGIFPSGRIADLTPDDIEQVLGVNFKGTVYIVQAALDALTAS-GHGRV
Mb0950c|M.bovis_AF2122/97 NNVGGTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGGGSV
Rv0927c|M.tuberculosis_H37Rv NNVGGTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGGGSV
MUL_4442|M.ulcerans_Agy99 NNVGGTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGGGGI
MSMEG_4330|M.smegmatis_MC2_155 NTVGKVLRKPIAETTEAEYDSMFDINSKAAYFFLQQAGKRLNDN---GSI
TH_3122|M.thermoresistible__bu NTVGRVLRKPIVETTEAEYDAMFDINAKAAYFFLQEAGKRLNDG---GSV
MAB_4657c|M.abscessus_ATCC_199 NTVGKVIKKPFTEITEEEYDSASAVNAKSAFLFLKEAGKHVNDN---GKI
Mvan_5843|M.vanbaalenii_PYR-1 NNSAVAHIAPIDDFPAEEFDRLVAINIGGVYWAVRSAIRYLSEG---ARI
. . : .: . :
MMAR_4033|M.marinum_M VNMSSVSG-KVGMVGQTNYSAAKAGIVGMTKAAAKELAHVGVRVNAIAPG
MAV_1572|M.avium_104 INMSSVSG-KVGMVGQTNYSAAKAGIVGMTKAAAKELAYLGVRVNAIAPG
MLBr_02565|M.leprae_Br4923 IVLSSMAG-IAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPG
Mflv_1235|M.gilvum_PYR-GCK IITSSITGPITGFPGWSHYGASKAAQLGFLRTAAIELAPKKITVNAVLPG
Mb0950c|M.bovis_AF2122/97 INISSTMG-RLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIAPG
Rv0927c|M.tuberculosis_H37Rv INISSTMG-RLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIAPG
MUL_4442|M.ulcerans_Agy99 INITSTMG-RVAGRGFAAYGTAKVALAHYTRLAALDLCPR-IRVNAIAPG
MSMEG_4330|M.smegmatis_MC2_155 TTIVTSLLAAYT-DGYSTYAGSKSPVEHFTRAASKEFGVRGINVNNIAPG
TH_3122|M.thermoresistible__bu VTILTALLAAFT-DGYSTYAGAKSPVEHFTRAAAKEFAPRQISVNNVAPG
MAB_4657c|M.abscessus_ATCC_199 VTLVTSLLGAYT-PFYAAYAGTKAPVEHFTRAASKEYGERGISVTAVGPG
Mvan_5843|M.vanbaalenii_PYR-1 INIGSINADRIPGPGLSVYGMTKGAVSSFTRGLARDLGPRGITVNNVQPG
: : *. :* : : : :. : **
MMAR_4033|M.marinum_M LIRSAMTEAMPQ--RIWDQKLAEVPMG----RAGEPSEVASVALFLACDL
MAV_1572|M.avium_104 LIRSAMTEAMPQ--RIWDSKVAEVPMG----RAGEPSEVASVALFLASDM
MLBr_02565|M.leprae_Br4923 FIETQMTAAIPL--ATREVGRRLNSLL----QGGLPVDVAETIAYFAIPA
Mflv_1235|M.gilvum_PYR-GCK NILTEGLVEMGQ--SYIDQMAAAVPAG----KLGEVADIGNAALFFATDE
Mb0950c|M.bovis_AF2122/97 SILTSALEVVAANDELRAPMEQATPLR----RLGDPVDIAAAAVYLASPA
Rv0927c|M.tuberculosis_H37Rv SILTSALEVVAANDELRAPMEQATPLR----RLGDPVDIAAAAVYLASPA
MUL_4442|M.ulcerans_Agy99 SILTSALEVVAANDELRAPMEQATPLR----RLGDPVDIAAAAVYLASPA
MSMEG_4330|M.smegmatis_MC2_155 PMDTPFFYPEETPERVEFHKSQAMGN-----RLTDIGDIAPLAVFLAD-E
TH_3122|M.thermoresistible__bu PMDTPFFYGQETPERVTFHKSQGLGN-----RLTRIEDIAPLVAFLTD-E
MAB_4657c|M.abscessus_ATCC_199 PMDTPFFYPAEGEDAVAYHKTAAALSPFSKTGLTDIEDIAPFIRFLVS-D
Mvan_5843|M.vanbaalenii_PYR-1 PIDTDAN-PDDGAFAESLKQVVAQGR------YGHTSDVASLVSFLAGPE
: : ::. ::.
MMAR_4033|M.marinum_M SSYMTGTVLDVTGGRFI----------
MAV_1572|M.avium_104 SSYMTGTVMEITGGRHL----------
MLBr_02565|M.leprae_Br4923 SNAVTGNVIRVCGQAMLGA--------
Mflv_1235|M.gilvum_PYR-GCK AAYITGQSLVVDGGQILPESPEALADL
Mb0950c|M.bovis_AF2122/97 GSFLTGKTLEVDGGLTFPNLDLPIPDL
Rv0927c|M.tuberculosis_H37Rv GSFLTGKTLEVDGGLTFPNLDLPIPDL
MUL_4442|M.ulcerans_Agy99 GSFLTGKTLEVDGGLTHPNLDIPVPDL
MSMEG_4330|M.smegmatis_MC2_155 GHWINGQTLFTNGGYTTR---------
TH_3122|M.thermoresistible__bu GHWITGQTIFANGGYTTR---------
MAB_4657c|M.abscessus_ATCC_199 GWWITGQTILINGGYTTK---------
Mvan_5843|M.vanbaalenii_PYR-1 SGYITGAHLNVDGGFTV----------
. :.* : *