For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLPDIDSLALFVQAAEQRSLTKAAEACNIGVAGASRRIMLLEARLKTTLFDRTPRGMKLTPAGKELHERA KTVLAQVNEMQALMKGFGADSQRALRVLVSSSAILGSFPTDLAAFCHANTDVELSVEEHRGADIVRALVD CEADLGLIVDGTPTEGLETFAYERDRLAVLAPSRHAVASMAELSFEGLLDHRLISLENGSSIMRVLSEQA SIAGRAVRPTVQVRGFEGLCRLVQEGLGVGVLPLAVAQRVGNRPGLVMRPLPESWAELGFLIAVRPDRRR NATLSTLLEFLSGARLNAREPGDRRVQLVPCS
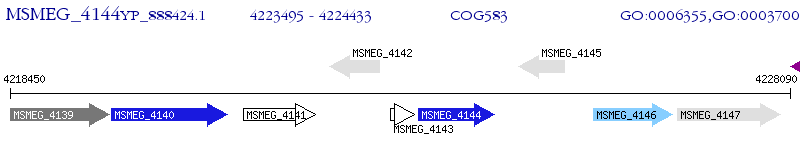
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4144 | - | - | 100% (312) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_0708 | - | 2e-19 | 28.57% (308) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_4609 | - | 2e-18 | 28.09% (299) | transcriptional regulator LysR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2303c | - | 3e-13 | 28.02% (207) | LysR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3565 | - | 2e-14 | 27.20% (250) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2282c | - | 3e-13 | 28.02% (207) | LysR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02663 | oxyS | 8e-08 | 28.85% (156) | putative LysR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2238 | - | 6e-18 | 27.24% (257) | LysR family transcriptional regulator |
| M. marinum M | MMAR_3419 | - | 7e-12 | 27.93% (179) | LysR family transcriptional regulator |
| M. avium 104 | MAV_1538 | - | 2e-15 | 24.79% (234) | transcriptional regulator, LysR family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2698 | - | 6e-12 | 27.93% (179) | LysR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3362 | - | 1e-16 | 27.49% (251) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2303c|M.bovis_AF2122/97 MPLSSRMPGLTCFEIFLAIAEAGSLGGAARELGLTQQAVSRRLASMEAQI
Rv2282c|M.tuberculosis_H37Rv MPLSSRMPGLTCFEIFLAIAEAGSLGGAARELGLTQQAVSRRLASMEAQI
MMAR_3419|M.marinum_M MPLSPRMPELGSFEIFLAIAQTGSLGAAARELGLTQQAVSKRLAAMEAHT
MUL_2698|M.ulcerans_Agy99 MPLSPRMPELASFEIFLAIAQTGSLGAAARELGLTQQAVSKRLAAMEAHT
Mflv_3565|M.gilvum_PYR-GCK -------MELRQLEYFVAIVEEGGFTRAASRERVAQPAVSAQIRRLERSV
Mvan_3362|M.vanbaalenii_PYR-1 -------MELRQLEYLVAVVEERSFTKAAQRERVAQPAVSAQIRRLERQV
MAB_2238|M.abscessus_ATCC_1997 -------MEIHQLRYLLAVADTSSFTKAAAALHVAQPGVSAQIRLLEREL
MLBr_02663|M.leprae_Br4923 -------MLFRQLEYFVAVAQERHFARAAERCYVSQPALSSAIAKLEREL
MAV_1538|M.avium_104 -------MAVGDYEWFITLAELQHVTAAAEQLHVAQPTLTRMLARLEQRL
MSMEG_4144|M.smegmatis_MC2_155 -----MLPDIDSLALFVQAAEQRSLTKAAEACNIGVAGASRRIMLLEARL
. :: .: . ** : : : :*
Mb2303c|M.bovis_AF2122/97 GVRLAIRTTRGSQLTPAGIVVAEWAARLLEVADEIDAGLGSLRTEGRQRI
Rv2282c|M.tuberculosis_H37Rv GVRLAIRTTRGSQLTPAGIVVAEWAARLLEVADEIDAGLGSLRTEGRQRI
MMAR_3419|M.marinum_M GVTLAVRTTRGSQLTPAGLIVSEWAARLLEVAQEIDAGLGSLRKEGRERI
MUL_2698|M.ulcerans_Agy99 GVTLAVRTTRGSQLTPAGLIVSEWAARLLEVAQEIDAGLGSLRKEGRERI
Mflv_3565|M.gilvum_PYR-GCK GQPLLTRTSREVRLTRAGEALLPHARAALAAVRDGRAAVDDVAG------
Mvan_3362|M.vanbaalenii_PYR-1 GQPLLTRSHRGVRVTPAGAALLPHARAALAAVRDAQAAVDDVAN------
MAB_2238|M.abscessus_ATCC_1997 GQDLFDRSGRQVRLTDAGKAVIPYARAALAAVDGVREAADEIGG------
MLBr_02663|M.leprae_Br4923 NVTLINRGHSFEGLTPEGERLVVWAKRILAEHDAFKAEVHAVRSG-----
MAV_1538|M.avium_104 GVALFDRHGRRLSLNTYGRIFYEHARRAQLELDSARRAIADLADP-----
MSMEG_4144|M.smegmatis_MC2_155 KTTLFDRTPRGMKLTPAGKELHERAKTVLAQVNEMQALMKGFGAD-----
* * :. * . * .
Mb2303c|M.bovis_AF2122/97 RVVASQTIAEQLMP-HWMLSLRAADMRRGGTVPEVILTATN-SEHAIAAV
Rv2282c|M.tuberculosis_H37Rv RVVASQTIAEQLMP-HWMLSLRAADMRRGGTVPEVILTATN-SEHAIAAV
MMAR_3419|M.marinum_M RVAASQTIAEQLMP-HWLLALQDAAKRRGTTAPQVILTATN-SDHAIAAV
MUL_2698|M.ulcerans_Agy99 RVAASQTIAEQLMP-HWLLALQDAAKRRGTTAPQVILTATN-SDHAIAAV
Mflv_3565|M.gilvum_PYR-GCK LLRGSVAIGTVTLH-PMNVAHLIAEFHADHPDVEITLGTAT-SDVLIAEL
Mvan_3362|M.vanbaalenii_PYR-1 LVRGAVAIGTVTLH-PVDVARLMADFHAEHPGVEITLGTDT-SDVLLAKL
MAB_2238|M.abscessus_ATCC_1997 LIRGHIAFGTVTSH-GADLSSLLSAFHRDHPAISITLTEAG-CEELVAGL
MLBr_02663|M.leprae_Br4923 -ITGTLRLGTVPTA-STTASLVLSAFCSEHPLIKVQICSRLPVTELHRRL
MAV_1538|M.avium_104 -AVGEIRLAYLSSFGSTVVPRLIARFKESSPRVTFTLEEGA-AESIADRV
MSMEG_4144|M.smegmatis_MC2_155 -SQRALRVLVSSSAILGSFPTDLAAFCHANTDVELSVEEHR-GADIVRAL
. . : . . : :
Mb2303c|M.bovis_AF2122/97 RDGIADLGFIENPCPP--TGLGSVVVARDELVVVVPPGHKWARRSRVVSA
Rv2282c|M.tuberculosis_H37Rv RDGIADLGFIENPCPP--TGLGSVVVARDELVVVVPPGHKWARRSRVVSA
MMAR_3419|M.marinum_M RDGTADLGFVENPGPP--TGLGSRVVGHDDLVVVVPPGHKWTRRPSPVTA
MUL_2698|M.ulcerans_Agy99 RDGTADLGFVENPGPP--TGLGSRVVGHDDLVVVVPPGHKWTRRPSPVTA
Mflv_3565|M.gilvum_PYR-GCK GDGRLDLAIVSIAVDEDLPGLEVAVITDEAVEAAVADGHPLAHR-RSLSL
Mvan_3362|M.vanbaalenii_PYR-1 EDGRLDAAIVSIGADEDPAGLEYEVITDEALEAAVTAQHRLARR-KSLSL
MAB_2238|M.abscessus_ATCC_1997 REGRLDGAIVALGAQR-LAGLGLQVIDDQPLVAAVAADDLLATR-RTITL
MLBr_02663|M.leprae_Br4923 GDFELDAVIVHSAPDD--THAVDLVPLYHEHYVLVSPADMLPPGASVLAW
MAV_1538|M.avium_104 RSGGVDIGVVSPRPVER--TLAWRSLFRQRLGVAVPDGHRFARG-AAVSM
MSMEG_4144|M.smegmatis_MC2_155 VDCEADLGLIVDGTPT--EGLETFAYERDRLAVLAPSRHAVASM-AELSF
. * .: . . .. . . ::
Mb2303c|M.bovis_AF2122/97 RELAQTPLVTREPNSGIRDSLTAALRDTLGEDMQQAPPVLELSSAAAVRA
Rv2282c|M.tuberculosis_H37Rv RELAQTPLVTREPNSGIRDSLTAALRDTLGEDMQQAPPVLELSSAAAVRA
MMAR_3419|M.marinum_M RELAETPLVTREPRSGIRDSLTVALRQVLGEDMVQASPVLELSSAAAMRA
MUL_2698|M.ulcerans_Agy99 RELAEMPLVTREPRSGIRDSLTVALRQVLGEDMVQASPVLELSSAAAMRA
Mflv_3565|M.gilvum_PYR-GCK EQLSRHPLISLPAGTGLRTRLDTACAAAG----LRPRIAFEATSPAELAE
Mvan_3362|M.vanbaalenii_PYR-1 KQLCEYPVISLPAGTGLRSRLDDACAAAG----LRPRIAFEATSPLELAD
MAB_2238|M.abscessus_ATCC_1997 QRLCEHTLICLPTGSGVRSILESECRSLG----LSPNVAFEASSPRALVS
MLBr_02663|M.leprae_Br4923 PEASQLPLALLTPDMRDRQLIDRAFAGHA----ITATPQVETDSVASLFA
MAV_1538|M.avium_104 VDLADEPFVTMHPGFGMRRLLDELCAAAQ----FRPRVALQAPNLTTAAS
MSMEG_4144|M.smegmatis_MC2_155 EGLLDHRLISLENGSSIMRVLSEQASIAG----RAVRPTVQVRGFEGLCR
. : .: .
Mb2303c|M.bovis_AF2122/97 AVLAGAGPAAMSRLAIADDLAFGRLLAVDIPALNLRRQLRAIWVGGR--T
Rv2282c|M.tuberculosis_H37Rv AVLAGAGPAAMSRLAIADDLAFGRLLAVDIPALNLRRQLRAIWVGGR--T
MMAR_3419|M.marinum_M AVLAGAGPAAMSRLAIADDLAVGRLRAISIPGLDLRRQLRAIWVGGR--V
MUL_2698|M.ulcerans_Agy99 AVLAGAGPAAMSRLAIADDLAVGRLRAISIPGLDLRRQLRAIWVGGR--V
Mflv_3565|M.gilvum_PYR-GCK LARHGLGVAILPQSMARGGSGLHALRLA----PELRGRLVWAWRADG-AN
Mvan_3362|M.vanbaalenii_PYR-1 LARHGLGVAILPRSMARGSAGLHPLRLS----PELRGRLVWAWRTDI-SG
MAB_2238|M.abscessus_ATCC_1997 LVEQGLGIAVVPAPYVRNRAGVLALDIR----PALRGRLALAWRLDGPVG
MLBr_02663|M.leprae_Br4923 QVATGNWACIVPHTWLWTTPIGAEIRAVEMVDPVLKADVALATNSAGPGS
MAV_1538|M.avium_104 LVAAGLGISLVP---IDGSSYPSGVSVKPLADADAYRDVGMIWDSGRPLP
MSMEG_4144|M.smegmatis_MC2_155 LVQEGLGVGVLPLAVAQRVGNRPGLVMRPLPESWAELGFLIAVRPDRRRN
. * :. : .
Mb2303c|M.bovis_AF2122/97 PPAGAIRDLLSHITSRST-------------
Rv2282c|M.tuberculosis_H37Rv PPAGAIRDLLSHITSRST-------------
MMAR_3419|M.marinum_M PPAGAIRELLSHITSGGE-------------
MUL_2698|M.ulcerans_Agy99 PPAGAIRELLSHITSGGE-------------
Mflv_3565|M.gilvum_PYR-GCK PAARVLGERARAMAVSPGSGAPAVVPGA---
Mvan_3362|M.vanbaalenii_PYR-1 PAARLLNARARAMIAATG--TPAVIPGA---
MAB_2238|M.abscessus_ATCC_1997 PAARAFLERARAVL-----------------
MLBr_02663|M.leprae_Br4923 PVARALVASAKQLALNEFFEAQLAGITRRR-
MAV_1538|M.avium_104 RSARDFIAGAAAVGHGGW-------------
MSMEG_4144|M.smegmatis_MC2_155 ATLSTLLEFLSGARLNAREPGDRRVQLVPCS
: