For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPRKPWSGVIPDEDLASFSREFDNEDRPLSAGTKPALLIIDMTRAFVDSTYPTGWSPTGAPAVQANAELL SAARRAHLPIFFTRGYPGPDDPATPSERGRWKRAANPRPLPPGTPPGDVIAEPLTPHAGEVVIDKGAKPS GFFGTALSSYLIYEGCDTVIVTGMTTSGCVRATVLDAFQYNFHVVVPHECCADRSQISHKVSLFDMHMKY GDVIDLSETIAYIESLSSQQHALSAS
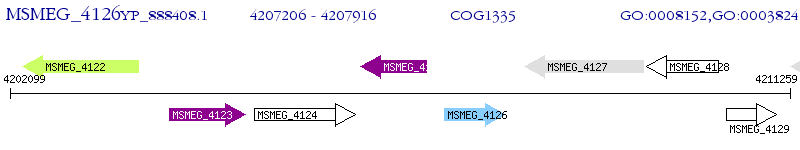
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4126 | - | - | 100% (236) | hydrolase, isochorismatase family protein |
| M. smegmatis MC2 155 | MSMEG_4379 | - | 7e-08 | 26.09% (161) | isochorismatase hydrolase |
| M. smegmatis MC2 155 | MSMEG_2243 | - | 2e-07 | 29.37% (126) | isochorismatase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2069c | pncA | 2e-06 | 33.55% (155) | pyrazinamidase/nicotinamidas PNCA (PZase) |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2043c | pncA | 2e-05 | 32.90% (155) | pyrazinamidase/nicotinamidas PNCA (PZase) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3431c | - | 2e-10 | 28.65% (171) | isochorismatase hydrolase |
| M. marinum M | MMAR_2657 | - | 3e-08 | 30.72% (153) | isochorismatase family protein |
| M. avium 104 | MAV_5193 | - | 5e-09 | 30.77% (182) | isochorismatase family protein |
| M. thermoresistible (build 8) | TH_2198 | - | 8e-05 | 34.25% (73) | isochorismatase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3053 | - | 5e-13 | 31.96% (219) | isochorismatase hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3431c|M.abscessus_ATCC_199 -----------------------MTGQVDAQPYPWPFDGPVDPGRT----
MMAR_2657|M.marinum_M --------------------------------------------------
Mvan_3053|M.vanbaalenii_PYR-1 -----------------------MNHTVDVPAEPSSFP--LVAGRT----
Mb2069c|M.bovis_AF2122/97 --------------------------------------------MR----
Rv2043c|M.tuberculosis_H37Rv --------------------------------------------MR----
MSMEG_4126|M.smegmatis_MC2_155 ---------------MPRKPWSGVIPDEDLASFSREFDNEDRPLSAGTKP
MAV_5193|M.avium_104 MLSADHRFQQTMTVRFGRRGRVPITGWVQSRRGVGMAGEDHYTRPCADSA
TH_2198|M.thermoresistible__bu ------------------------------------------MSDT----
MAB_3431c|M.abscessus_ATCC_199 AVLCIDWQVDFCGRGGYVDTMGYDLSLTRAGLEPTARVLAAARA-VGMTI
MMAR_2657|M.marinum_M --------------------MGYDLSLVRAPLGPAQTVLEAARK-LGLLI
Mvan_3053|M.vanbaalenii_PYR-1 ALIVIDMQRDFLLPGGFGESLGNDVGRLLKVVPPLASLIAAARA-AGVMV
Mb2069c|M.bovis_AF2122/97 ALIIVDVQNDFCEGGSLAVTGG-------AALARAISDYLAEAA-DYHHV
Rv2043c|M.tuberculosis_H37Rv ALIIVDVQNDFCEGGSLAVTGG-------AALARAISDYLAEAA-DYHHV
MSMEG_4126|M.smegmatis_MC2_155 ALLIIDMTRAFVDSTYPTGWSPTG----APAVQANAELLSAARR-AHLPI
MAV_5193|M.avium_104 ALVLIDVQRDFYADDAPMRVEGTS-----AALGAMAELARAFRR-RELPI
TH_2198|M.thermoresistible__bu AVVVVDMFNTYDHEDAELLTPN-----VESILDPLAKLIAEARHRDDVDL
: :
MAB_3431c|M.abscessus_ATCC_199 IHTREGHRPDLSDLPANKRWRSARIGAEIGVAGPCGRILVKGEPGWEIVP
MMAR_2657|M.marinum_M VHTREGHRPDLSDLPANKRWRSARIGAEIGVAGPCGRVLTRGEPGWEIIP
Mvan_3053|M.vanbaalenii_PYR-1 VHTREGHQPDLSDCPPAKLNRGTPS-KRIGDPGRYGRILIRGEYGHDIID
Mb2069c|M.bovis_AF2122/97 VATKDFHIDPGDDFSGTPDYSSSWPPHCVSGTPGADFHPSLDTSAIEAVF
Rv2043c|M.tuberculosis_H37Rv VATKDFHIDPGDHFSGTPDYSSSWPPHCVSGTPGADFHPSLDTSAIEAVF
MSMEG_4126|M.smegmatis_MC2_155 FFTRGYPGPDDPATPSERGRWKRAANPRPLPPG--------TPPGDVIAE
MAV_5193|M.avium_104 VHVVRLYRADGSNADPVRRRFIEDGARVAEPGSPGSQIAPELLPKAVELD
TH_2198|M.thermoresistible__bu IYVNDNHGDFTAEFSDVVR-------------------TALDGARPDLVR
. .
MAB_3431c|M.abscessus_ATCC_199 EVAPLPGEPIIDKPGK--------GAFYATDLDLVLRTRGIRYIVLTGIT
MMAR_2657|M.marinum_M EMEPLPGEMVVDKLGK--------GSFYATDLELILTTRRITHLIFTGIA
Mvan_3053|M.vanbaalenii_PYR-1 ELAPIDGEIVIDKPGK--------GAFYATSLSDVLTEAGITQLLITGVT
Mb2069c|M.bovis_AF2122/97 YKGAYTG--AYSGFEG--------VDENGTPLLNWLRQRGVDEVDVVGIA
Rv2043c|M.tuberculosis_H37Rv YKGAYTG--AYSGFEG--------VDENGTPLLNWLRQRGVDEVDVVGIA
MSMEG_4126|M.smegmatis_MC2_155 PLTPHAGEVVIDKGAKP-------SGFFGTALSSYLIYEGCDTVIVTGMT
MAV_5193|M.avium_104 HQLLLSGGFQQIGPAEHVMYKPRWGAFYGTKLEQHLRESGTDTLVFAGCN
TH_2198|M.thermoresistible__bu PIVPDDGCRALLKVRH--------SVFYATALEYLLTRLSARRVILTGQV
* .* * * : ..*
MAB_3431c|M.abscessus_ATCC_199 TDVCVHTTMREANDRGFECLILSDCTGATDAGNHAAALKMVTMQGGVFGA
MMAR_2657|M.marinum_M TDVCVHTTMREANDRGYECLLLSDCTGATDYANHLAALKMITMQGGVFGA
Mvan_3053|M.vanbaalenii_PYR-1 TEVCVHTTTREANDRGYQCLVVSDCVGSYFPEFQRVGLEMIKAQGGIFGW
Mb2069c|M.bovis_AF2122/97 TDHCVRQTAEDAVRNGLATRVLVDLTAGVSADTTVAALEEMRTASVELVC
Rv2043c|M.tuberculosis_H37Rv TDHCVRQTAEDAVRNGLATRVLVDLTAGVSADTTVAALEEMRTASVELVC
MSMEG_4126|M.smegmatis_MC2_155 TSGCVRATVLDAFQYNFHVVVPHECCADRSQISHKVSLFDMHMKYGDVID
MAV_5193|M.avium_104 FPNCPRTSIYEASERDFRIVLVTDAISG----LYDRGAQECRAIGVAVRD
TH_2198|M.thermoresistible__bu TEQCILYSALDAYVRHFEVVVPPDAVAHIDAGLGEAALQMMARN--MSAE
* : :* : : . .
MAB_3431c|M.abscessus_ATCC_199 VADSNQLLGAL----------
MMAR_2657|M.marinum_M HATNGALLEAFNKVRDRRG--
Mvan_3053|M.vanbaalenii_PYR-1 VADTSSVIPALARLSSIPA--
Mb2069c|M.bovis_AF2122/97 SS-------------------
Rv2043c|M.tuberculosis_H37Rv SS-------------------
MSMEG_4126|M.smegmatis_MC2_155 LSETIAYIESLSSQQHALSAS
MAV_5193|M.avium_104 TAQTLDWLGG-----------
TH_2198|M.thermoresistible__bu LPPSADCLP------------
.