For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IEKQTLPLDGIRVVAIEQAVAAPLCSRHLADLGADVIKIERPGGGDFARDYDTYAKGMSSHFVWLNRGKR SVALDLKDDAGREVLLRLLDTADVFLTNLAPGAVDRIVDDSTLAERNPRLVWCAISGYGPDGPYEKRKAF DLLVQGEAGVTLNTGEPGCPAKPGVSLADLGGGIYAVTAILSALRQRDATGRGDRVHVAMFDVLVEWMSP LLIAYLEAGVEIPPAGTRHATITPYGPFTTQDGKVINIAVQNDRQWLALCGCVGASALSDDPDLRHNAGR LARRNDVEKAVASAVAGISEKALVERLDAAGVPWGRMNGVADVVAHPQLQAGNRWTTATLPSGATVPVVA APFRFASRPTAADDSASVPAVGAHTDELLEELGIR*
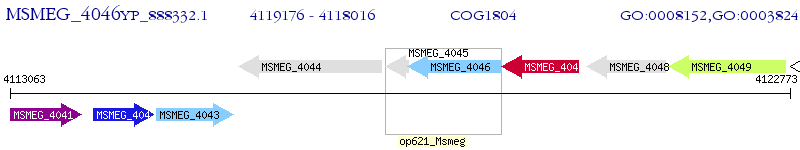
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4046 | - | - | 100% (386) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. smegmatis MC2 155 | MSMEG_1992 | - | 1e-78 | 42.19% (384) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. smegmatis MC2 155 | MSMEG_4834 | - | 1e-75 | 40.05% (387) | L-carnitine dehydratase/bile acid-inducible protein F |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3300 | - | 1e-41 | 32.64% (386) | hypothetical protein Mb3300 |
| M. gilvum PYR-GCK | Mflv_2443 | - | 6e-80 | 41.65% (389) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. tuberculosis H37Rv | Rv3272 | - | 3e-42 | 32.90% (386) | hypothetical protein Rv3272 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4651c | - | 9e-43 | 33.23% (328) | putative L-carnitine dehydratase |
| M. marinum M | MMAR_1267 | - | 3e-43 | 33.59% (393) | acyl-CoA transferase |
| M. avium 104 | MAV_2651 | - | 2e-84 | 45.97% (372) | L-carnitine dehydratase/bile acid-inducible protein F |
| M. thermoresistible (build 8) | TH_2496 | - | 1e-75 | 41.65% (389) | PUTATIVE L-carnitine dehydratase/bile acid-inducible protein F |
| M. ulcerans Agy99 | MUL_2621 | - | 2e-44 | 34.10% (393) | acyl-CoA transferase |
| M. vanbaalenii PYR-1 | Mvan_4208 | - | 2e-78 | 41.39% (389) | L-carnitine dehydratase/bile acid-inducible protein F |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2443|M.gilvum_PYR-GCK -MTASQ-SAGPLAGVTVVAMEQAVAAPMCTRVLADFGARIIKVENPNGGD
Mvan_4208|M.vanbaalenii_PYR-1 -MSSSQPSAAPLAGVTVVAMEQAVAAPMCTRVLADFGARIIKVENPNGGD
TH_2496|M.thermoresistible__bu MAESEHADPAPLAGITVVALEQAVSAPMCTRVLADFGARVIKVENPDGGD
MAV_2651|M.avium_104 -----------------MSLEQAVAAPYATRQLADLGARVIKVERPQGGD
MSMEG_4046|M.smegmatis_MC2_155 ---MIEKQTLPLDGIRVVAIEQAVAAPLCSRHLADLGADVIKIERPGGGD
Mb3300|M.bovis_AF2122/97 --MPTSNPAKPLDGFRVLDFTQNVAGPLAGQVLVDLGAEVIKVEAP-GGE
Rv3272|M.tuberculosis_H37Rv --MPTSNPAKPLDGFRVLDFTQNVAGPLAGQVLVDLGAEVIKVEAP-GGE
MMAR_1267|M.marinum_M --MPEGNPAKPLDGFRVLDFTQNVAGPLAGQVLADLGAEVIKVEAP-GGE
MUL_2621|M.ulcerans_Agy99 --MPEGNPAKPLDGFRVLDFTQNVAGPLAGQVLADLGAEVIKVEAP-GGE
MAB_4651c|M.abscessus_ATCC_199 -------MTGALDGIRVLELGTLIAGPFAGRLLGDMGAEVLKIEPPGAPD
: : ::.* . : * *:** ::*:* * . :
Mflv_2443|M.gilvum_PYR-GCK FARAYDDVVNGPGGLAAHFVWSNRGKESVTLNTKSPAGLEVLHRLLDRAD
Mvan_4208|M.vanbaalenii_PYR-1 FARDYDDVVNGPGGLAAHFVWSNRGKESVTLNTKTAGGLDLLHRLLDRAD
TH_2496|M.thermoresistible__bu FARHYDDVVKG---QAAHFVWANRGKESVTLNLKSEAGMAVLHRLLDEAD
MAV_2651|M.avium_104 FARGYDRTVHG---ESSYFVWLNRGKESLTVDLKQPEGRRIVERLLEGAD
MSMEG_4046|M.smegmatis_MC2_155 FARDYDTYAKG---MSSHFVWLNRGKRSVALDLKDDAGREVLLRLLDTAD
Mb3300|M.bovis_AF2122/97 AARQITSVLPGRPPLATYFLPNNRGKKSVTVDLTTEQAKQQMLRLADTAD
Rv3272|M.tuberculosis_H37Rv AARQITSVLPGRPPLATYFLPNNRGKKSVTVDLTTEQAKQQMLRLADTAD
MMAR_1267|M.marinum_M AARHITAVLPHRPPLATYFLPNNRGKKSVSVDLSTDTARRQILRLADTAD
MUL_2621|M.ulcerans_Agy99 AARHITAVLPHRPPLATYFLPNNRGKKSVSVDLSTDTARRQILRLADTAD
MAB_4651c|M.abscessus_ATCC_199 PLRTWGQAEVDG--HHVFWTVHARNKKAITLDLRTEAGRALFLDLVDKSD
* .: *.*.::::: . . * : :*
Mflv_2443|M.gilvum_PYR-GCK AFVSNLAPGATARLGLSPADLAERHPQVIPVEIDGYGPGGPISHKRAYDL
Mvan_4208|M.vanbaalenii_PYR-1 AFVSNLAPGATARLGVAPADLAARHPRVIPVEIDGYGPGGPLSHKRAYDL
TH_2496|M.thermoresistible__bu VLVSNLAPGALARMGLAADDLKTSHPRVLPVQIDGYGAGGPISHKRAYDL
MAV_2651|M.avium_104 VLVQNLAPGAAARMGLDTESVARAHPRTITATISGYGQDGPWRDRKAYDL
MSMEG_4046|M.smegmatis_MC2_155 VFLTNLAPGAVDRI-VDDSTLAERNPRLVWCAISGYGPDGPYEKRKAFDL
Mb3300|M.bovis_AF2122/97 VVLEAFRPGTMEKLGLGPDDLRSRNPNLIYARLTAYGGNGPHGSRPGIDL
Rv3272|M.tuberculosis_H37Rv VVLEAFRPGTMEKLGLGPDDLRSRNPNLIYARLTAYGGNGPHGSRPGIDL
MMAR_1267|M.marinum_M VVLEGFRPGVMERMGLGPEDLRARNPKLIYARLSAFGGNGPHGTRPGVDL
MUL_2621|M.ulcerans_Agy99 VVLEGFRPGVMERMGLGPEDLRARNPKLIYARLSAFGGNGPHGTRPGVDL
MAB_4651c|M.abscessus_ATCC_199 VIVENFRPGTLERWGLGYDVLAQRNKGIILVRVSGYGQTGPDAHKAGYAS
..: : **. : : : : : : .:* ** : .
Mflv_2443|M.gilvum_PYR-GCK LVQAESGSCAVTGYP-GMPAKPGPAVADFTSGLYAAMSIVSLLYGRATR-
Mvan_4208|M.vanbaalenii_PYR-1 LVQAESGSCAVTGYP-GMPAKPGPAVADFSTGLYAAISILALLYGRARH-
TH_2496|M.thermoresistible__bu LIQAESGACAVTGYP-GMPAKPGPPVADISTGLYAALSVMALLYGRDRRD
MAV_2651|M.avium_104 LVQAEAGLLSVTGSP-AEVARTGVSIADIAAGMFAFSGILTALYTRATT-
MSMEG_4046|M.smegmatis_MC2_155 LVQGEAGVTLNTGEP-GCPAKPGVSLADLGGGIYAVTAILSALRQRDAT-
Mb3300|M.bovis_AF2122/97 VVAAEAGMTTGMPTPEGKPQIIPFQLVDNASGHVLAQAVLAALLHRERN-
Rv3272|M.tuberculosis_H37Rv VVAAEAGMTTGMPTPEGKPQIIPFQLVDNASGHVLAQAVLAALLHRERN-
MMAR_1267|M.marinum_M MVAAEAGMTTGMPTPDGKPQIIPFQLVDNASGHVLAQAVLAALLNRERH-
MUL_2621|M.ulcerans_Agy99 MVAAEAGMTTGMPTPDGKPQIIPFQLVDNASGHVLAQAVLAALLNRERH-
MAB_4651c|M.abscessus_ATCC_199 VAEAASGLRHLNGFPGGPPPRMALSIGDTLAGMFAAQGALAALYRRTVT-
: . :* * . : * * . :: * *
Mflv_2443|M.gilvum_PYR-GCK -PEPGPAPHVELSLFDVMTDVMGYALTYTQHSG-------IDQEPLGVGS
Mvan_4208|M.vanbaalenii_PYR-1 -PGSAPAPSVELSLFDVMTDVMGYALTYTQHSG-------IDQEPLGVGS
TH_2496|M.thermoresistible__bu RATPGPVPAVAVSLFDTMTDIMGYQLTYTQHSG-------VDQQPLGMSS
MAV_2651|M.avium_104 ----GAVRPVSVSLFDALVEWMSQPLYYGRYGG-------KAPPRTGARH
MSMEG_4046|M.smegmatis_MC2_155 ----GRGDRVHVAMFDVLVEWMSPLLIAYLEAG-------VEIPPAGTRH
Mb3300|M.bovis_AF2122/97 ----GVADVVQVAMYDVAVGLQANQLMMHLNRAASDQPK-PEPAPKAKRR
Rv3272|M.tuberculosis_H37Rv ----GVADVVQVAMYDVAVGLQANQLMMHLNRAASDQPK-PEPAPKAKRR
MMAR_1267|M.marinum_M ----GVADTVRVAMYDVAVGLQANQLTMHLNRPSSDAPKRPEQTPAGARR
MUL_2621|M.ulcerans_Agy99 ----GVADTVRVAMYDVAVGLQANQLTMHLNRPSSDAPKRPEQTPAGARR
MAB_4651c|M.abscessus_ATCC_199 ----GRGQVVDAALTESCLAVQESTIPDYDVGR-------VVRGPSGTRL
. * :: : : .
Mflv_2443|M.gilvum_PYR-GCK P----AVAPYGAFPTRDGQTVVLGTTNDREWQRVAREIIDRPDLADDPRF
Mvan_4208|M.vanbaalenii_PYR-1 P----AVAPYGAFPTRDGQTVVLGTTNDREWQRVAREIIDRPDLADDPRF
TH_2496|M.thermoresistible__bu P----AVAPYGAFRTRDGQTVVLGTTNDREWQRLARDIIERPDLADDPAY
MAV_2651|M.avium_104 P----TIAPYGPVTAGDGGTVLLGVQNSAEWLRFCEIVLERRDLVDEPRF
MSMEG_4046|M.smegmatis_MC2_155 A----TITPYGPFTTQDGKVINIAVQNDRQWLALCGCVG-ASALSDDPDL
Mb3300|M.bovis_AF2122/97 KGVGFATQPSDAFRTADG-YIVISAYVPKHWQKLCYLIG-RPDLVEDQRF
Rv3272|M.tuberculosis_H37Rv KGVGFATQPSDAFRTADG-YIVISAYVPKHWQKLCYLIG-RPDLVEDQRF
MMAR_1267|M.marinum_M KRVAFATQPSDAFRAADG-YLLISAYVPKHWQRLCEIIG-RPDLLDDERF
MUL_2621|M.ulcerans_Agy99 KRVAFATQPSDAFRAADG-YLLISAYVPKHWQRLCEIIG-QPDLLDDERF
MAB_4651c|M.abscessus_ATCC_199 EG----IAPSNIYQSADGSWVVIAANQDTVFRRLCEAMG-RPELSSDARF
* . : ** : :.. : .. : * .:
Mflv_2443|M.gilvum_PYR-GCK ATNPGRCANRDVLEEAIGAWCARHDLAEIQDKADRAGIGNSRYNLPSEVI
Mvan_4208|M.vanbaalenii_PYR-1 ATNSDRCAHRQILEEAIGSWCAQHDLAEIQKTADAAGIGNSRYNLPSEVV
TH_2496|M.thermoresistible__bu ATNSDRVRHRDVLNEAIAAWCARHDLAHIQKTADDAGIGNSRYNLPSEVV
MAV_2651|M.avium_104 ATNPDRVAHRDELESIITTVAARLSAEELEARLQSAGVAHARMNGVDHLW
MSMEG_4046|M.smegmatis_MC2_155 RHNAGRLARRNDVEKAVASAVAGISEKALVERLDAAGVPWGRMNGVADVV
Mb3300|M.bovis_AF2122/97 AEQRSRSINYAELTAELELALASKTATEWVQLLQANGLMACLAHTWKQVV
Rv3272|M.tuberculosis_H37Rv AEQRSRSINYAELTAELELALASKTATEWVQLLQANGLMACLAHTWKQVV
MMAR_1267|M.marinum_M IDQRARAINYAELTEELESALAAKTAAQWVELLHEGGLMACLAYTWKQVV
MUL_2621|M.ulcerans_Agy99 IDQRARAINYAELTEELESALAAKTAAQWVELLHEGGLMACLAYTWKQVV
MAB_4651c|M.abscessus_ATCC_199 ANHVARGRNQDDLDAIIAQWAAQRLPTDIIDVLSAAGVIAGPINTVADVV
: * . : : * *: .:
Mflv_2443|M.gilvum_PYR-GCK AHPHLVARDRWRTVATPKGDIQALRPPPVISDFEQPMG----AIPGLGEH
Mvan_4208|M.vanbaalenii_PYR-1 VHPHLAARDRWRTVATPKGDIQALRPPPVISDFEQPMG----AIPGLGEH
TH_2496|M.thermoresistible__bu AHPHLSARDRWRPVDTPNGQITALRPPPVIEGFEQPMG----PVPGLGQH
MAV_2651|M.avium_104 LHPVLVGRDRWHEIQTPGGPAAALPPPAGLAGVDPVIG----DVPALGEH
MSMEG_4046|M.smegmatis_MC2_155 AHPQLQAGNRWTTATLPSGATVPVVAAPFRFASRPTAADDSASVPAVGAH
Mb3300|M.bovis_AF2122/97 DTPLFAESDLTLEVGRGADTITVIRTPARYASFRAVVTD---PPPTAGEH
Rv3272|M.tuberculosis_H37Rv DTPLFAENDLTLEVGRGADTITVIRTPARYASFRAVVTD---PPPTAGEH
MMAR_1267|M.marinum_M TTPLFAENELALQVGSGDDAVTVIRTPARYSSFSAVLAE---PPAATGEH
MUL_2621|M.ulcerans_Agy99 TTPLFAENELALQVDSGDDAVTVIRTPARYSSFSAVLAD---PPPATGEH
MAB_4651c|M.abscessus_ATCC_199 QDPQLKAREMLVEHFDERLGRSVLGPGVVPVLSESPGGVRNAGPARPGQD
* : : : . . * .
Mflv_2443|M.gilvum_PYR-GCK TDAVLT-EFGVSAEELAQLRAEGAIGPAYRG
Mvan_4208|M.vanbaalenii_PYR-1 TDAVLT-EIGVTPDELRQLRADGAIGPAYRG
TH_2496|M.thermoresistible__bu TDAVLSGLLGMTDDEIAALREQGAIGPAYN-
MAV_2651|M.avium_104 SRKILH-ELGYSAQATDELIAAGVTTA----
MSMEG_4046|M.smegmatis_MC2_155 TDELLE-ELGIR-------------------
Mb3300|M.bovis_AF2122/97 NAVFLARP-----------------------
Rv3272|M.tuberculosis_H37Rv NAVFLARP-----------------------
MMAR_1267|M.marinum_M NETLLGES-----------------------
MUL_2621|M.ulcerans_Agy99 NETLLGES-----------------------
MAB_4651c|M.abscessus_ATCC_199 NDDVYVGLLGKSAQDIERLRAEGVL------
. .