For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHTTIPSTMDGVYLPGDSTAVLKQFDVRPPGPGQVLLEMGASGICGSDIGYIYRGYKGYRGIDGPAYQG VVAGHEPAGRIVATGEGVTRFGVGDRVLLYHIVGCGLCDNCRRGFYISCSGDRASYGWQRDGGHARYVLA EERTCIPLPDELSFVDGALIACGFATAYEGLRRAGVSGDHDLLVVGLGPVGLAAGMIGRGMGAATVIGVE PSTTRREWADSLGIFDATIPAGPDALDTITELTRGAGASVTIDCSGSRAGRSLALEAAAEWGNISLVGEG NDLHTEVSDTLLHKQLTIHASWVTSLPTMSELATNLVRWNLKPETVVSDIFPLSQAARAYELAAGTSRGK VCLVPDGNAA
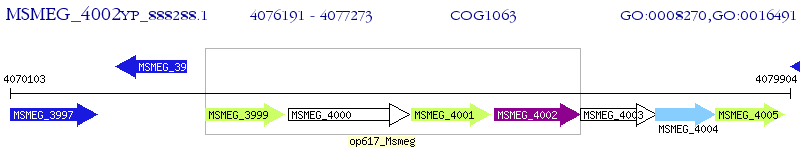
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4002 | - | - | 100% (360) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_3605 | - | 1e-29 | 28.25% (354) | sorbitol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3094 | - | 2e-29 | 28.77% (365) | oxidoreductase, zinc-binding dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0784c | adhB | 4e-19 | 28.03% (289) | zinc-containing alcohol dehydrogenase NAD dependant ADHB |
| M. gilvum PYR-GCK | Mflv_4517 | - | 1e-33 | 33.43% (338) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 6e-32 | 31.59% (345) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 9e-20 | 27.30% (282) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_2084 | - | 9e-27 | 30.87% (298) | zinc-type alcohol dehydrogenase AdhD |
| M. marinum M | MMAR_2790 | - | 1e-28 | 30.67% (326) | dehydrogenase |
| M. avium 104 | MAV_2804 | - | 4e-27 | 29.39% (330) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4349 | - | 3e-31 | 30.75% (335) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_2962 | - | 2e-28 | 30.67% (326) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0527 | - | 1e-32 | 33.33% (249) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
TH_4349|M.thermoresistible__bu --------MKALRLMDWKSEPELVEVPDPTPGPGQVLVKIGGAGACHSDL
MMAR_2790|M.marinum_M --------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVAVSAAGICGSDL
MUL_2962|M.ulcerans_Agy99 --------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDL
Rv1895|M.tuberculosis_H37Rv --------MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGSDL
MAV_2804|M.avium_104 --------MRTVVVDGPRSIRVDTRPDPVLPGPDGAIVEVSAAGICGSDL
Mflv_4517|M.gilvum_PYR-GCK --------MRTVVVDSQRSIRVDTRPDPRLPGPDGVIVDVTASAICGSDL
Mb0784c|M.bovis_AF2122/97 ------MKTKGALIWEFNQPWSVEEIEIGDPRKDEVKIQMEAAGMCRSDH
MAB_2084|M.abscessus_ATCC_1997 ------MKTRAAVLWGLEQKWEVEEVDLDPPGPGEVLVRLAATGLCHSDE
MLBr_01784|M.leprae_Br4923 ----MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDL
Mvan_0527|M.vanbaalenii_PYR-1 --------MRAAIYHGREDVR-IEELPDPSPRAGEVVIEVARAGICGTDL
MSMEG_4002|M.smegmatis_MC2_155 MSHTTIPSTMDGVYLPGDSTAVLKQFDVRPPGPGQVLLEMGASGICGSDI
. * . . : : . * :*
TH_4349|M.thermoresistible__bu HLMHDFEPGMM----PWQLPFTLGHENAGWVAAVGAGVGTVTEGDAVAVY
MMAR_2790|M.marinum_M HFYEGEYP--------LAEPVALGHEAVGTVVEKGPDVHTVEVGDQVMVS
MUL_2962|M.ulcerans_Agy99 HFYEGEYP--------LAEPVALGHEAVGTVVEKGPDVHTVEVGDQVMVS
Rv1895|M.tuberculosis_H37Rv HFYEGEYP--------FTEPVALGHEAVGTIVEAGPQVRTVGVGDLVMVS
MAV_2804|M.avium_104 HFYEADFP--------MPDPIALGHEAIGTVVEAGPQVRNVKVGDQVMVS
Mflv_4517|M.gilvum_PYR-GCK HFLDGHYP--------IDQPVPIGHEAVGVVVETGSDVTRFTNGDRVLVS
Mb0784c|M.bovis_AF2122/97 HLVTGDIPMAG-------FPVLGGHEGAGIVTEVGPGVDDFAPGDHVVLA
MAB_2084|M.abscessus_ATCC_1997 HLVTGDLPIP--------LPVVGGHEGAGTVVECGEGVEDLSEGDSVILT
MLBr_01784|M.leprae_Br4923 TYREGGIN--------DRYPFLLGHEAAGTVEVVGPGVTAVEPGDFVILN
Mvan_0527|M.vanbaalenii_PYR-1 HEYIAGPMH-------AAPGVVIGHEYSGTVVGVGSGVREFTEGDRVCGV
MSMEG_4002|M.smegmatis_MC2_155 GYIYRGYKGYRGIDGPAYQGVVAGHEPAGRIVATGEGVTRFGVGDRVLLY
. *** * : * * . ** *
TH_4349|M.thermoresistible__bu GPWGCGTCERCMGGAENYCENPA----GAPVLSGGAGLGMDGGMAEYLLV
MMAR_2790|M.marinum_M SVAGCGACAGCATRDPAMCVS------GPQIFGSGA---LGGAQADLLAV
MUL_2962|M.ulcerans_Agy99 SVAGCGACAGCATRDPAMCVS------GPQIFGSGA---LGGAQADLLAV
Rv1895|M.tuberculosis_H37Rv SVAGCGVCPGCETHDPVMCFS------GPMIFGAGV---LGGAQADLLAV
MAV_2804|M.avium_104 SVAGCGSCAGCATRDPVLCHS------GFQIFGGGV---LGGAQADLLAV
Mflv_4517|M.gilvum_PYR-GCK SVTGCGRCAGCLTLDPVKCVH------GPQIFGTGL---LGGAQSDLLAV
Mb0784c|M.bovis_AF2122/97 FIPSCGKCPSCQAGMRNLCDLGAGLLAGESVTDGSFRIQARGQNVYPMTL
MAB_2084|M.abscessus_ATCC_1997 FLPSCGRCSYCARGMGNLCELGAALMMGP-QIDGTYRFHARGEDVGQMCL
MLBr_01784|M.leprae_Br4923 WRAVCGQCRACKRGRPSYCFDTFNAQQKMTLIDGTELTPALGIGAFADKT
Mvan_0527|M.vanbaalenii_PYR-1 GVFGCGECGFCKQGAEALCGA-----------VGFIGFAVNGALARYASL
MSMEG_4002|M.smegmatis_MC2_155 HIVGCGLCDNCRRGFYISCSG----------DRASYGWQRDGGHARYVLA
** * * * *
TH_4349|M.thermoresistible__bu P---------AERLVVKLPESMTPVTAAALTDAGLTPYHAIRRSWPKLTP
MMAR_2790|M.marinum_M PA--------ADFQVLKIPAAITTEQALLLTDNLATGWAAARR--ADILF
MUL_2962|M.ulcerans_Agy99 PA--------TDFQVLKIPAAITTEQALLLTDNLATGWAAARR--ADIPF
Rv1895|M.tuberculosis_H37Rv PA--------ADFQVLKIPEGITTEQALLLTDNLATGWAAAQR--ADISF
MAV_2804|M.avium_104 PA--------ADFQLLKIPEGISTEQALLLTDNLATGWAAAQR--ADIPF
Mflv_4517|M.gilvum_PYR-GCK PA--------ADFQLLAVPDGIGTEEALLLTDNLPTGWAAAKR--ADIPV
Mb0784c|M.bovis_AF2122/97 LGTFSPYMVVHRSSVVKIDPSVPFEVACLVGCGVTTGYGSAVR-TADVRP
MAB_2084|M.abscessus_ATCC_1997 LGTFSEYTVVPQASVVKIDGGIPLDRAALIGCGVTTGYGSAVR-TGEVRA
MLBr_01784|M.leprae_Br4923 LV--------HSGQCTKVDPAADPAVAGLLGCGVMAGLGAAIN-TAAVSR
Mvan_0527|M.vanbaalenii_PYR-1 PT----------KALFRIPDEISLAEAAVVEP-IASAYHAVRR--SGLAA
MSMEG_4002|M.smegmatis_MC2_155 EER----------TCIPLPDELSFVDGALIACGFATAYEGLRR--AGVSG
: . : : . . :
TH_4349|M.thermoresistible__bu TSTAVVIGVGGLGHVAVQLVKATTAARLIAVDNRSEALELAREMGADHVV
MMAR_2790|M.marinum_M GATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRASEWGATAVA
MUL_2962|M.ulcerans_Agy99 GATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRAAEWGATAVA
Rv1895|M.tuberculosis_H37Rv GSAVAVIGLGAVGLCALRSAFIHGAATVFAVDRVKGRLQRAATWGATPIP
MAV_2804|M.avium_104 GGTVAVIGLGAVGLCALRSALFQGAATVFAVDQVDGRLARAARWGATPLK
Mflv_4517|M.gilvum_PYR-GCK GGTVAVIGAGAVGQCALRSALALGAARVFAVDPVVARRERAAAAGATPMD
Mb0784c|M.bovis_AF2122/97 GDDVAIVGLGGVGMAALQGAVSAGARYVFAVEPVEWKRDQALKFGATHVY
MAB_2084|M.abscessus_ATCC_1997 GDTVVVVGAGGIGMNAIQGARIAGALAIVAVDPVNFKREQASGFGATHTA
MLBr_01784|M.leprae_Br4923 DDTVAVIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWARTFGATHTV
Mvan_0527|M.vanbaalenii_PYR-1 GGTVFIAGAGPIGLALVQFSLAQGATQVIVSEVSATRRVAAHRVGATRVI
MSMEG_4002|M.smegmatis_MC2_155 DHDLLVVGLGPVGLAAGMIGRGMGAATVIGVEPSTTRREWADSLGIFDAT
: * * :* * :. : * *
TH_4349|M.thermoresistible__bu RSDE-QAVEAIWELTGGRGADVLLDFVG--ADATIELARAAARPLGDVTI
MMAR_2790|M.marinum_M SP----AAEAIVAATGGRGADSVIDAVG--TDASMTDALNAVRAGGTVSV
MUL_2962|M.ulcerans_Agy99 SP----AAEAIVAATSGRGADSVIDAVD--TDASMTDALNAVRAGGTVSV
Rv1895|M.tuberculosis_H37Rv SP----AAETILAATRGRGADSVIDAVG--TDASMSDALNAVRPGGTVSV
MAV_2804|M.avium_104 AP----ALEAILAATGGRGADAVIDAVA--TDASLTEALNAVRPGGTVSV
Mflv_4517|M.gilvum_PYR-GCK SP----AAQTILEATDGLGADSVIDAVG--TDASIDDALAGVRTGGTVSV
Mb0784c|M.bovis_AF2122/97 PDIN-AALMGIAEVTYGLMAQKVIITVGKLDGADVDSYLTITAKGGTCVL
MAB_2084|M.abscessus_ATCC_1997 ASMD-EAWSLVSDMTRGKLADVCILTTDVAEGSYTAEALALVGKRGRVVV
MLBr_01784|M.leprae_Br4923 NALELEVVKTIQDLTSGFGVDVVIDTVG--RPETWKQAFYARDLAGTVVL
Mvan_0527|M.vanbaalenii_PYR-1 DPLAEDAVEVVRMLTNGNGVDISFDAAG--VQPALDAALGVLRPRGRLMV
MSMEG_4002|M.smegmatis_MC2_155 IPAGPDALDTITELTRGAGASVTIDCSG--SRAGRSLALEAAAEWGNISL
. : * * .. : * :
TH_4349|M.thermoresistible__bu VGIAGGS----VPLSFFSQPYEVSIQTTYWG------TRPELVELLDLAA
MMAR_2790|M.marinum_M VGVHDLQPFPLPALTCLLRSITLRLTIAPVQR-----TWSELIPLLESGR
MUL_2962|M.ulcerans_Agy99 VGVHDLQPFPLPALTCLLRSITLRLTIAPVQR-----TWSELIPLLESGR
Rv1895|M.tuberculosis_H37Rv VGVHDLQPFPVPALTCLLRSITLRMTMAPVQR-----TWPELIPLLQSGR
MAV_2804|M.avium_104 VGVHDMNPFPLNALGCLIRSITLRMTTAPVQR-----TWPELIPLLQSGR
Mflv_4517|M.gilvum_PYR-GCK VGVHDLTPYPLPALVCLLRSLTLRMTTAPVQQ-----TWAELVPLLQAGR
Mb0784c|M.bovis_AF2122/97 TAIGSLVDTQVTLNLAMLTLLQKNIQGTIFGGGNPHYDIPKLLSMYKAGK
MAB_2084|M.abscessus_ATCC_1997 TAIGHPDESSISGSLLEMTLYEKQIRGALYGSSNAPHDIPRLVELYSAGL
MLBr_01784|M.leprae_Br4923 VGVPTPDMRLDMPLLDFFS-HGGSLKSSWYGDCLPERDFPTLVDLYLQGR
Mvan_0527|M.vanbaalenii_PYR-1 VAIWEAPAGIDINRSVMREANIGFSFCYEAQR-----QVPAILDLLATGA
MSMEG_4002|M.smegmatis_MC2_155 VGEGNDLHTEVSDTLLHKQLTIHASWVTSLPT------MSELATNLVRWN
.. . :
TH_4349|M.thermoresistible__bu RGLVHVESTTYPLDDAL-QAYRDLRAGRVRGRAVIVP-------------
MMAR_2790|M.marinum_M LDVDGIFTTSLPLDEAA-KGYSIAGSRSGNDVKVLLKV------------
MUL_2962|M.ulcerans_Agy99 LDVDGIFTTSLPLDEAA-KGYSIAGSRSGNDVKVLLKV------------
Rv1895|M.tuberculosis_H37Rv LDVDGIFTTTLPLDEAA-KGYATARARSGEELRFCLRPDSRDVLGAHETV
MAV_2804|M.avium_104 LDVDGIFTTTMPLDEAA-IGYATAGSRSGDDVKILLKP------------
Mflv_4517|M.gilvum_PYR-GCK LSVDGIFTDAMPLAEAE-AAYAAAFSRSGEHLKVHLVP------------
Mb0784c|M.bovis_AF2122/97 LNLDDMVTTAYKLEQIN-DGYQDMLNGKNIRGVIRYTDDDR---------
MAB_2084|M.abscessus_ATCC_1997 LKLDELVTREYSLDQIN-EGYQDMRSGRNIRGLVRF--------------
MLBr_01784|M.leprae_Br4923 LPLGKFVSERIGLGDVE-EAFHKIHGGKVLRSVVML--------------
Mvan_0527|M.vanbaalenii_PYR-1 INLGELITDEIPLDAVVSQGLEELRVNRDAHVKILIDPSA----------
MSMEG_4002|M.smegmatis_MC2_155 LKPETVVSDIFPLSQAA-RAYELAAGTSRGKVCLVPDGNAA---------
. : * . .
TH_4349|M.thermoresistible__bu -------------------------------
MMAR_2790|M.marinum_M -------------------------------
MUL_2962|M.ulcerans_Agy99 -------------------------------
Rv1895|M.tuberculosis_H37Rv DLYVHVRRCQSVADLQLEGAADGVDGPSMLN
MAV_2804|M.avium_104 -------------------------------
Mflv_4517|M.gilvum_PYR-GCK -------------------------------
Mb0784c|M.bovis_AF2122/97 -------------------------------
MAB_2084|M.abscessus_ATCC_1997 -------------------------------
MLBr_01784|M.leprae_Br4923 -------------------------------
Mvan_0527|M.vanbaalenii_PYR-1 -------------------------------
MSMEG_4002|M.smegmatis_MC2_155 -------------------------------