For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAHETRTWFITGSSRGFGRALVKAALDAGDNVAATARKPGQLADFAETYGDRVLALPLDVTDPSAARTAV ATAREAFGRIDIVVNNAGYANVAPIETGDDEDFRTQFETNFWGVYNVSKAALPVLREQRGGLIIQFSSMG GRVGGSPGIGSYQAAKFAIDGFSRVLLAETAPFGVKVLVVEPSGFATDWAGASMEVHDIPADYADTVGGM SAIRQSDSVTAGDPVRAAEILVQMSRRDDIPYHLPIGVHSVEGCLQQDEFLLGEDRKWAAVGRSADFSEA YPVAFPPDTVSSSA
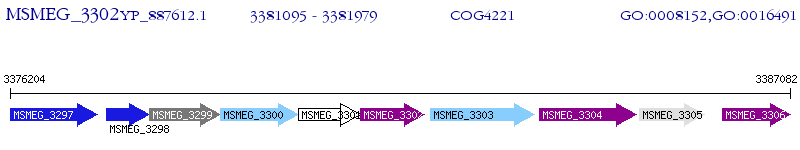
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3302 | - | - | 100% (294) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_1114 | - | 7e-54 | 45.75% (247) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2026 | - | 1e-51 | 42.55% (275) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 5e-24 | 36.36% (198) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 5e-63 | 43.82% (283) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 5e-24 | 36.36% (198) | oxidoreductase |
| M. leprae Br4923 | MLBr_01094 | - | 7e-19 | 34.57% (188) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3919c | - | 5e-23 | 36.08% (194) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_1614 | - | 4e-45 | 42.68% (246) | short chain dehydrogenase |
| M. avium 104 | MAV_4982 | - | 4e-81 | 53.90% (282) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_3670 | - | 2e-53 | 45.27% (243) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 2e-45 | 42.68% (246) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0982 | - | 1e-53 | 42.96% (270) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3302|M.smegmatis_MC2_155 ---------MAHETRTWFITGSSRGFGRALVKAALDAGDNVAATARKPGQ
MAV_4982|M.avium_104 ------------MPKTWFITGSSRGLGRALTTAVLDHGDHVVATARRPAQ
Mflv_0295|M.gilvum_PYR-GCK ------------MSKVLLISGASRGLGRAITESALAAGHLVVAGVRATSS
TH_3670|M.thermoresistible__bu MGSNVWG------SKVWFITGASRGFGREWAIAALDRGDRVAATARDVST
Mvan_0982|M.vanbaalenii_PYR-1 MPGHPTGIDPGMTEKVWFITGASRGFGREWAIAALDRGDKVAATARDLST
MMAR_1614|M.marinum_M -------------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARSVGG
MUL_3423|M.ulcerans_Agy99 -------------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARSVGG
Mb1079|M.bovis_AF2122/97 ------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGA
Rv1050|M.tuberculosis_H37Rv ------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGA
MLBr_01094|M.leprae_Br4923 --------MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEG
MAB_3919c|M.abscessus_ATCC_199 --------MKNFDNKVAVITGAGSGIGRSLALALAQRGARLALSDIDTAG
. .::*.. *:*. . * .
MSMEG_3302|M.smegmatis_MC2_155 LADFA---ETYGDRVLALPLDVTDPSAARTAVATAREAFGRIDIVVNNAG
MAV_4982|M.avium_104 LDELV---DRYGPRIRACELDVTDRAAARRAIAAAVTAFGRIDVVVNNAG
Mflv_0295|M.gilvum_PYR-GCK LSDLA---TREPDRLAVVELDVTDDRHARAAVATAVERFGRLDVLVNNAG
TH_3670|M.thermoresistible__bu LDDLR---DRFGDAVLAMTLDVTDRAADFAAVQRAHDHFGRLDIVVNNAG
Mvan_0982|M.vanbaalenii_PYR-1 LDDLT---AKYGDALLPLTLDVTDRDADFAAVTQAHDHFGRLDIVVNNAG
MMAR_1614|M.marinum_M VADLV---QRSPERVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAG
MUL_3423|M.ulcerans_Agy99 VADLA---QRSPERVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAG
Mb1079|M.bovis_AF2122/97 LRRVAREIEAAGGRAMVAPLDVSSSESVRAMVADVVGEFGRIDVVFNNAG
Rv1050|M.tuberculosis_H37Rv LRRVAREIEAAGGRAMVAPLDVSSSESVRAMVADVVGEFGRIDVVFNNAG
MLBr_01094|M.leprae_Br4923 LAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAG
MAB_3919c|M.abscessus_ATCC_199 VADTAGRCEKAGATAIPFELDVADRAAVYAHAADVKNEFGQVNLVFNNAG
: **::. . ** :: : ****
MSMEG_3302|M.smegmatis_MC2_155 YANVAPIETGDDED-FRTQFETNFWGVYNVSKAALPVLREQRGGLIIQFS
MAV_4982|M.avium_104 YANSAPVEEVSDDD-FRAQVETNFFGVVNVTKAALPVLHRQRAGHIVQIS
Mflv_0295|M.gilvum_PYR-GCK YANMSAIEDVDVDD-FRAQIETNFFGVVNLTRAALPAMRRQRDGHIVQIS
TH_3670|M.thermoresistible__bu YGHFGFVEELTETE-VRDQMETNFFGALWVTQAALPYLRRQRSGHIIQVS
Mvan_0982|M.vanbaalenii_PYR-1 YGQFGFVEELSEQE-ARAQIDTNVFGALWITQAALPYLRAQRSGHLIQVS
MMAR_1614|M.marinum_M YGYRAAVEEGDDAE-VRDLFETHFFGTVALIKAVLPDMRARRSGAIVNIS
MUL_3423|M.ulcerans_Agy99 YGYRAAVEEGDDAE-VRDLFETHFFGTVALIKAVLPDMRARRSGAIVNIS
Mb1079|M.bovis_AF2122/97 VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSGRIMNMS
Rv1050|M.tuberculosis_H37Rv VSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLPIMKQQRSGRIMNMS
MLBr_01094|M.leprae_Br4923 IAFTGDVEVSHFKD-IERVMDVDYWGVVNGTKAFLPYLISSGDGHVINIS
MAB_3919c|M.abscessus_ATCC_199 VALSADVKDMEWDD-FDWLMNINFWGVAHGTKAFLPHLIESGDGHIVNVS
. . :. : .: . *. : ** : * :::.*
MSMEG_3302|M.smegmatis_MC2_155 SMGGRVGGSPGIGSYQAAKFAIDGFSRVLLAETAPFG--VKVLVVEPSGF
MAV_4982|M.avium_104 SVGGRVGGSPGIAAYQASKFAVAGFSEALAAEVAPLG--IKVTIAEPGAL
Mflv_0295|M.gilvum_PYR-GCK SVGGRLVR-PGLAAYQSAKWAVTGFSGVLAVEVAPLG--IKVTVLEPGGM
TH_3670|M.thermoresistible__bu SIGG-VSAFADLGAYHASKWALEGLSQSLSLEVAPFG--IHVTLIEPGGY
Mvan_0982|M.vanbaalenii_PYR-1 SIGG-IVAFPNLGIYHASKWALEGFSQALAQEVAEFG--IHVTLIEPGGY
MMAR_1614|M.marinum_M SIAVALTP-VGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAF
MUL_3423|M.ulcerans_Agy99 SIAVALTP-VGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAF
Mb1079|M.bovis_AF2122/97 SVVG-RKAFARFAGYSSAMHAIAGFSDALRQELRGSG--IAVSVIHPALT
Rv1050|M.tuberculosis_H37Rv SVVG-RKAFARFAGYSSAMHAIAGFSDALRQELRGSG--IAVSVIHPALT
MLBr_01094|M.leprae_Br4923 SVFG-LFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGI
MAB_3919c|M.abscessus_ATCC_199 SVFG-LMGIPSQSAYNAAKFAVRGFTESLRQEIRAAKFPVGVTCVHPGGI
*: . * :: *: *:: * * : * *.
MSMEG_3302|M.smegmatis_MC2_155 ATDWAGASMEVHD-IPADYADTVGGMSAIRQ-SDSVTAGDPVRAAEILVQ
MAV_4982|M.avium_104 RTEWAGSSMAVAT-VSPDYESTVGAMNRRRAGFNRNAPGDPARAAHAIIE
Mflv_0295|M.gilvum_PYR-GCK RTDWAGSSMRIAP-VREEYAATVGAAASLSD-PNVLGASDPAKVAGLLLD
TH_3670|M.thermoresistible__bu ATDWGGSSARRSA-TMPEYAESRAEADRVRS-QRVARPGDPKASAAALLK
Mvan_0982|M.vanbaalenii_PYR-1 STDWAGASAKRAT-ALPAYAEAHAATERTRQ-ARTARPGDPQASARALLT
MMAR_1614|M.marinum_M RTDFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIIT
MUL_3423|M.ulcerans_Agy99 RTDFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIIT
Mb1079|M.bovis_AF2122/97 QTPLLANVDPADMPPPFRSLTPIPVHWVAAAVLDGVARRRARVVVPFQPR
Rv1050|M.tuberculosis_H37Rv QTPLLANVDPADMPPPFRSLTPIPVHWVAAAVLDGVARRRARVVVPFQPR
MLBr_01094|M.leprae_Br4923 KTAIARNAT-AAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKAR
MAB_3919c|M.abscessus_ATCC_199 KTNIASSARGIPDGVDRETVRKGFQ-LMAITRPDSAARIILRGVEKNRPR
*
MSMEG_3302|M.smegmatis_MC2_155 MSRRDDIPYHLPIGVHSVEGCLQQDEFLLGEDRKWAAVGRSADFSEAYPV
MAV_4982|M.avium_104 VVIADDPPLRLLLGSDALSAALAKSHQQIAEANRWAELSKSIDYP---PV
Mflv_0295|M.gilvum_PYR-GCK VIGMPDPPRRLLVGPDAYRYATAAGRDLLAVDERWRTLSMSTAADDVSPE
TH_3670|M.thermoresistible__bu VVDAEKPPLRVFFGELPLQIVNADYQSRLQTWEEWQPIAVLAQG------
Mvan_0982|M.vanbaalenii_PYR-1 VVDAPEPPLRVFFGELPLELARADYESRLANWEKWQPVAIEAQG------
MMAR_1614|M.marinum_M AVESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP----
MUL_3423|M.ulcerans_Agy99 AVESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP----
Mb1079|M.bovis_AF2122/97 LLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPTESAKAQAA
Rv1050|M.tuberculosis_H37Rv LLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYRGSVYRHQPTESAKAQAA
MLBr_01094|M.leprae_Br4923 VLVGPDAKVANVVVRFSG-----------GAGYQRLFAQVASRLILNQR-
MAB_3919c|M.abscessus_ATCC_199 VLIGPDARVFDAIPRIIGPRYEDLMAPFYGMGRYGIGKKIAGRFGIEL--
MSMEG_3302|M.smegmatis_MC2_155 AFPPDTVSSSA
MAV_4982|M.avium_104 ATPHRAARSR-
Mflv_0295|M.gilvum_PYR-GCK QLDPLGSASR-
TH_3670|M.thermoresistible__bu -----------
Mvan_0982|M.vanbaalenii_PYR-1 -----------
MMAR_1614|M.marinum_M -----------
MUL_3423|M.ulcerans_Agy99 -----------
Mb1079|M.bovis_AF2122/97 QPERGYSSAR-
Rv1050|M.tuberculosis_H37Rv QPERGYSSAR-
MLBr_01094|M.leprae_Br4923 -----------
MAB_3919c|M.abscessus_ATCC_199 -----------