For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KNFDNKVAVITGAGSGIGRSLALALAQRGARLALSDIDTAGVADTAGRCEKAGATAIPFELDVADRAAVY AHAADVKNEFGQVNLVFNNAGVALSADVKDMEWDDFDWLMNINFWGVAHGTKAFLPHLIESGDGHIVNVS SVFGLMGIPSQSAYNAAKFAVRGFTESLRQEIRAAKFPVGVTCVHPGGIKTNIASSARGIPDGVDRETVR KGFQLMAITRPDSAARIILRGVEKNRPRVLIGPDARVFDAIPRIIGPRYEDLMAPFYGMGRYGIGKKIAG RFGIEL*
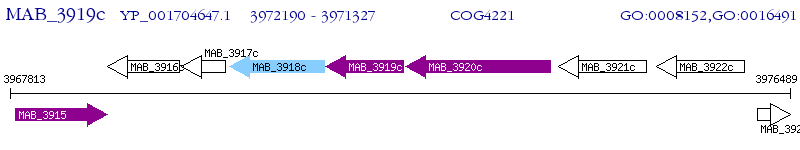
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3919c | - | - | 100% (287) | putative short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_1389c | - | 3e-80 | 54.37% (263) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_1388c | - | 7e-79 | 53.01% (266) | short-chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3112 | - | 2e-76 | 53.76% (266) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2216 | - | 8e-74 | 51.89% (264) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3085 | - | 2e-76 | 53.76% (266) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01094 | - | 5e-71 | 51.16% (258) | short chain alcohol dehydrogenase |
| M. marinum M | MMAR_4195 | - | 1e-73 | 52.96% (270) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_1384 | - | 3e-74 | 52.81% (267) | short chain alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5053 | - | 1e-75 | 53.38% (266) | short chain alcohol dehydrogenase |
| M. thermoresistible (build 8) | TH_1828 | - | 6e-75 | 52.24% (268) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_4501 | - | 7e-73 | 52.22% (270) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4479 | - | 5e-78 | 53.79% (264) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2216|M.gilvum_PYR-GCK MQGFAGKVAVVTGAGSGIGQALAIELGRSGALVAISDVDTEGLAVTEERL
Mvan_4479|M.vanbaalenii_PYR-1 MEGFAGKVAVVTGAGSGIGQALAIELGRSGARVAISDVDTEGLAVTEERL
MMAR_4195|M.marinum_M MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
MUL_4501|M.ulcerans_Agy99 MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
MLBr_01094|M.leprae_Br4923 MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTEGQL
MAV_1384|M.avium_104 MQGFAGKVAVVTGAGSGIGQALAVELARSGAKVAISDVDLEGLAHTEEQL
MSMEG_5053|M.smegmatis_MC2_155 MEGFAGKVAVVTGAGSGIGQALAIELGRSGAKLAICDVDTEGLAATEERL
TH_1828|M.thermoresistible__bu MQGFAGKVAVITGAGSGIGQALALELGRSGAKVAISDVNTEGLTETEDRL
Mb3112|M.bovis_AF2122/97 MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDGLAKTVRLA
Rv3085|M.tuberculosis_H37Rv MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDGLAKTVRLA
MAB_3919c|M.abscessus_ATCC_199 MKNFDNKVAVITGAGSGIGRSLALALAQRGARLALSDIDTAGVADTAGRC
*..* .****:********::**: *.. * :*:.*:: *:: *
Mflv_2216|M.gilvum_PYR-GCK KAIGTHVKSDRLNVTERERFLQYADDVAEHFGKVHQIYNNAGIAFTGDIE
Mvan_4479|M.vanbaalenii_PYR-1 KAIGAQVKSDRLDVTERETFLLYADDVAEHFGSVNQIYNNAGIAFTGDIE
MMAR_4195|M.marinum_M TALGAEVKTDRLDVTEREAFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
MUL_4501|M.ulcerans_Agy99 TALGAEVKTDRLDVTERETFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
MLBr_01094|M.leprae_Br4923 KAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTGDVE
MAV_1384|M.avium_104 KAIGAQYKADRLDVTEREAFLAYADAVKEHFGKVNQIYNNAGIAFTGDVE
MSMEG_5053|M.smegmatis_MC2_155 KAIGAPVKADRLDVTEREAFLLYADAVKEHYGTVNQIYNNAGIAFTGDVE
TH_1828|M.thermoresistible__bu KAIGAPVKADRLDVTERAAFEAYADAVVEHFGVVNQIYNNAGIAYTGDIE
Mb3112|M.bovis_AF2122/97 QALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAGIAYNGNVD
Rv3085|M.tuberculosis_H37Rv QALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAGIAYNGNVD
MAB_3919c|M.abscessus_ATCC_199 EKAGATAIPFELDVADRAAVYAHAADVKNEFGQVNLVFNNAGVALSADVK
*: . .*:*::* . :* * .:* *: ::****:. ..::.
Mflv_2216|M.gilvum_PYR-GCK VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
Mvan_4479|M.vanbaalenii_PYR-1 ITQFKDIERVMDVDYWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
MMAR_4195|M.marinum_M VCAFKDIDRVMDVDFGGVLNGTKAFLPYLIASGDGHVINVSSVFGLFSVS
MUL_4501|M.ulcerans_Agy99 VCAFKDIDRVMDVDFGGVLNGTKTFLPYLIASGDGHVINVSSVFGLFSVS
MLBr_01094|M.leprae_Br4923 VSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLFSVP
MAV_1384|M.avium_104 VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
MSMEG_5053|M.smegmatis_MC2_155 ISEFKDIERVMDVDYWGVVNGTKAFLPHLIASGDGHVINVSSVFGLFSVP
TH_1828|M.thermoresistible__bu VSHFKDIERVMDVDFWGVVNGTKVFLPHLIASGDGHVINISSVFGLFSVP
Mb3112|M.bovis_AF2122/97 KSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISSLFGLIAVP
Rv3085|M.tuberculosis_H37Rv KSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISSLFGLIAVP
MAB_3919c|M.abscessus_ATCC_199 DMEWDDFDWLMNINFWGVAHGTKAFLPHLIESGDGHIVNVSSVFGLMGIP
:.*:: :::::: ** :***.***::* *****::*:**:***:.:.
Mflv_2216|M.gilvum_PYR-GCK GQGAYNAAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNAG-
Mvan_4479|M.vanbaalenii_PYR-1 GQAAYNSAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNAS-
MMAR_4195|M.marinum_M GQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNAT-
MUL_4501|M.ulcerans_Agy99 GQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNAT-
MLBr_01094|M.leprae_Br4923 GQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIARNAT-
MAV_1384|M.avium_104 GQAAYNSAKFAVRGFTEALRQEMAAAGHPVAVTTVHPGGIKTAIARNAT-
MSMEG_5053|M.smegmatis_MC2_155 GQAAYNSAKFAVRGFTEALRQEMAIAKHPVKVTTVHPGGIKTAIARNAT-
TH_1828|M.thermoresistible__bu GQAAYNAAKFAVRGFTEALRQEMILAGHPVKVTTVHPGGIKTGIARNMT-
Mb3112|M.bovis_AF2122/97 GQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKTAVARNAT-
Rv3085|M.tuberculosis_H37Rv GQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKTAVARNAT-
MAB_3919c|M.abscessus_ATCC_199 SQSAYNAAKFAVRGFTESLRQEIRAAKFPVGVTCVHPGGIKTNIASSARG
.*.***:**********:**:*: * ** *: *:****** :* .
Mflv_2216|M.gilvum_PYR-GCK AAEGLDAEELAKTFDKKLASTTPEKAAQVILDGVRKNKARILIGNDAKMF
Mvan_4479|M.vanbaalenii_PYR-1 AAEGLDAEELAKAFDKKLASTTPEKAAKIILDGVRKNRARILVGNDAKVF
MMAR_4195|M.marinum_M AAEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKAL
MUL_4501|M.ulcerans_Agy99 AAEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKAL
MLBr_01094|M.leprae_Br4923 AAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGPDAKVA
MAV_1384|M.avium_104 AAEGLDQAELAKLFDKRLAKTTPQRAAQIILDAVRKKKARVLVGSDAKAL
MSMEG_5053|M.smegmatis_MC2_155 AAEGLDAKELAEAFDKKLANTTPQRAAVIILDGVRKNKARVLVGPDAKIL
TH_1828|M.thermoresistible__bu AAEGLDAQELARAFDTKLARTSPEKAAQIILDGVRKNKARVLVGTDAKIL
Mb3112|M.bovis_AF2122/97 VADGEDQQTFAEFFDRRLALHSPEMAAKTIVNGVAKGQARVVVGLEAKAV
Rv3085|M.tuberculosis_H37Rv VADGEDQQTFAEFFDRRLALHSPEMAAKTIVNGVAKGQARVVVGLEAKAV
MAB_3919c|M.abscessus_ATCC_199 IPDGVDRETVRKGFQ-LMAITRPDSAARIILRGVEKNRPRVLIGPDARVF
.:* * . *: :* *: ** *: .* * :.*:::* :*:
Mflv_2216|M.gilvum_PYR-GCK EIAIRALG-AGYQSVFPKV--VARLTPPAK---------
Mvan_4479|M.vanbaalenii_PYR-1 DVVVRVLG-AGYQSLFPKA--VARLTPPAK---------
MMAR_4195|M.marinum_M DIVVRLTG-SGYQRLFMPV--LGRLVPASHR--------
MUL_4501|M.ulcerans_Agy99 DIVVRLTG-SGYQRLFMPV--LGRLVPASHR--------
MLBr_01094|M.leprae_Br4923 NVVVRFSGGAGYQRLFAQV--ASRLILNQR---------
MAV_1384|M.avium_104 DILVRLTG-SGYQRLFGPV--MSRLLPN-----------
MSMEG_5053|M.smegmatis_MC2_155 DVIVRITG-SGYQRLFSAV--AAKAMPR-----------
TH_1828|M.thermoresistible__bu DAIVRLTG-SGYQRLFPKF--VARTMPR-----------
Mb3112|M.bovis_AF2122/97 DVLARIMG-SSYQRLVAAG--VAKFFPWAK---------
Rv3085|M.tuberculosis_H37Rv DVLARIMG-SSYQRLVAAG--VAKFFPWAK---------
MAB_3919c|M.abscessus_ATCC_199 DAIPRIIG-PRYEDLMAPFYGMGRYGIGKKIAGRFGIEL
: * * . *: :. .: