For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDAVNSGRAARRVRLTHHVVELDDGHQVGVSVGGVGVPMVFLHGLGLNRRVYIRLLSRVAGLGFRIVAID APGHGDTGDLPPDADGFSERTALVLRTMDALGVEKAVLAGHSMGGRTAIHLAAVAPQRVLAAILVDAAAG SSFDATISTVTRSPRQMLNSVLGAIYDMHSDPLHMQIGALNRYLRMVAAVTMGKSFASRGFTGAARAIMQ SGACAALLAAMREHRIPTIVLHGEHDSIVPFDNACDVADVVDATLYRVRGACHSWLIGNPRRGADSIRQL LHGELGDVLREAATTLGIDDWRDAEDWDRTLVTSAAQVRTLAGAGVVIGPDSRERVEMDLVRRPGPTRAE AVPETPERATA
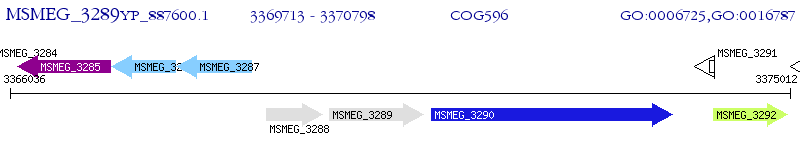
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3289 | - | - | 100% (361) | gp61 protein |
| M. smegmatis MC2 155 | MSMEG_6850 | - | 3e-11 | 36.97% (119) | alpha/beta hydrolase |
| M. smegmatis MC2 155 | MSMEG_1897 | pcaD | 3e-11 | 28.46% (260) | 3-oxoadipate enol-lactonase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3600c | bphD | 4e-10 | 25.19% (258) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. gilvum PYR-GCK | Mflv_3146 | - | 1e-11 | 26.02% (246) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3569c | bphD | 4e-10 | 25.19% (258) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. leprae Br4923 | MLBr_02603 | - | 5e-07 | 25.00% (228) | hypothetical protein MLBr_02603 |
| M. abscessus ATCC 19977 | MAB_1875c | - | 2e-10 | 26.67% (240) | putative hydrolase (alpha/beta fold) |
| M. marinum M | MMAR_1120 | - | 2e-10 | 29.23% (284) | hydrolase |
| M. avium 104 | MAV_4371 | - | 9e-11 | 26.50% (283) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_3727 | - | 1e-09 | 24.29% (247) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | MUL_0880 | - | 1e-10 | 29.23% (284) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4414 | - | 5e-17 | 27.71% (249) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3600c|M.bovis_AF2122/97 --------------------------------------------MTATEE
Rv3569c|M.tuberculosis_H37Rv --------------------------------------------MTATEE
Mvan_4414|M.vanbaalenii_PYR-1 ----------------------------------------------MSSE
Mflv_3146|M.gilvum_PYR-GCK --------------------------------------------------
TH_3727|M.thermoresistible__bu --------------------------------------------------
MMAR_1120|M.marinum_M MSAEETRRRQRAWLAGGAGLTAVATIVGASARRSLAERGSVCDDPYAGED
MUL_0880|M.ulcerans_Agy99 MSAEETRRRQRAWLAGGAGLTAVATIVGASARRSLAERGSVCDDPYAGED
MAV_4371|M.avium_104 -------------------MTAVATIVGASARRSMTQR-ATIEDPYADED
MSMEG_3289|M.smegmatis_MC2_155 ------------------------------------------MDAVNSGR
MAB_1875c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02603|M.leprae_Br4923 --------------------------------------------------
Mb3600c|M.bovis_AF2122/97 LTFESTSRFAEVDVDGPLKLHYHEAG--VGNDQTVVLLHGGGPGAASWTN
Rv3569c|M.tuberculosis_H37Rv LTFESTSRFAEVDVDGPLKLHYHEAG--VGNDQTVVLLHGGGPGAASWTN
Mvan_4414|M.vanbaalenii_PYR-1 ITEQYTSRDIETKSG---TIHYNEAG--EG--YPIVMLHGSGPGATGWSN
Mflv_3146|M.gilvum_PYR-GCK --------MPTITTQDGVEIFYKDWG----SGQPIVFSHGWPLSADDWDA
TH_3727|M.thermoresistible__bu --------MATITTSDGVEIFYRDWG----SGQPIVFSHGWPLTADDWDP
MMAR_1120|M.marinum_M FERLDSDRARVVTTPDGVPLAVREAGP-VDAPLTMVFAHGFCLRMGAFHF
MUL_0880|M.ulcerans_Agy99 FERLDSDRARVVTTPDGVPLAVREAGP-VDAPLTMVFAHGFCLRMGAFHF
MAV_4371|M.avium_104 FNTFDGDRALVVTTPDGVPLAVREAGP-PDAPLTMVFVHGFCLQMGAFHF
MSMEG_3289|M.smegmatis_MC2_155 AARRVRLTHHVVELDDGHQVGVSVGG----VGVPMVFLHGLGLNRRVYIR
MAB_1875c|M.abscessus_ATCC_199 ----MSEHRRTSISVDGHRLAYRQFG---SGDRVVVLSHAFLTSHYLEHA
MLBr_02603|M.leprae_Br4923 --MTITRTERNFYGVGGVRIVYDVWMPDTRPRAVIILAHGFGEHARRYDH
: ::: *.
Mb3600c|M.bovis_AF2122/97 FSRNIAV-LARHFHVLAVDQPG-YGHSDKRAEHGQFNRYAAMALKGLFDQ
Rv3569c|M.tuberculosis_H37Rv FSRNIAV-LARHFHVLAVDQPG-YGHSDKRAEHGQFNRYAAMALKGLFDQ
Mvan_4414|M.vanbaalenii_PYR-1 FGSNIKG-LADRFRVIAADMPG-WGQSE--AVKPDARDHVAAALQ-LLDG
Mflv_3146|M.gilvum_PYR-GCK QLMFF---LGHGYRVVAHDRRG-HGRSG-QVDDGHDMDHYADDLAAVVEH
TH_3727|M.thermoresistible__bu QMLFF---LNAGYRVIAHDRRG-HGRSS-PVPDGHDMDHYADDLAALVEH
MMAR_1120|M.marinum_M QRMRLGEQWGSRVRMVFYDQRG-HGQSGEADPQTYTLTQLGQDLETVLQA
MUL_0880|M.ulcerans_Agy99 ERMRLGEQWGSRVRMVFYDQRG-HGQSGEADPQTYTLTQLGQDLETVLQA
MAV_4371|M.avium_104 QRTRLPEQLGPDVRMVFYDQRG-HGRSGEAAPESYTLTQLGRDLQTVLQV
MSMEG_3289|M.smegmatis_MC2_155 LLSRVAG---LGFRIVAIDAPG-HGDTGDLPPDADGFSERTALVLRTMDA
MAB_1875c|M.abscessus_ATCC_199 WAQELAA---RGFRVICLDLLGTTADERPLEPDRYNSQALGRQLIGALDA
MLBr_02603|M.leprae_Br4923 VAHYFAA---AGLATYALDLRG-HGRSAGKRVLVRDLSEYNADFDILVGI
. * * . . .
Mb3600c|M.bovis_AF2122/97 LGLGR----VPLVGNSLGGGTAVRFALDYPARAGRLVLMGPGGLSINLFA
Rv3569c|M.tuberculosis_H37Rv LGLGR----VPLVGNSLGGGTAVRFALDYPARAGRLVLMGPGGLSINLFA
Mvan_4414|M.vanbaalenii_PYR-1 LGIEK----AAFVGNSMGGGTAIQFAVAHPDRISHLITMGSRAPGPTIIG
Mflv_3146|M.gilvum_PYR-GCK LDLHD----AVHVGHSTGGGEVVRYLARHGEDRAVKAALISAVPPLMVQT
TH_3727|M.thermoresistible__bu LGLRD----AVHVGHSTGGGVVVRYLARHGEERTAKAVLIAAVPPLMVRT
MMAR_1120|M.marinum_M TAPRGP---IVLVGHSMGGMTVLSHARQFPQRYGRRIVGAAVISSAAEGV
MUL_0880|M.ulcerans_Agy99 TAPRGP---IVLVGHSMGGMTVLSHARQFPQRYGRRIVGAAVISSAAEGV
MAV_4371|M.avium_104 VAPRGL---VVLVGHSMGGMTVLSHARQFPEQYGRRIVGAALISSAAEGV
MSMEG_3289|M.smegmatis_MC2_155 LGVEK----AVLAGHSMGGRTAIHLAAVAPQRVLAAILVDAAAGSSFDAT
MAB_1875c|M.abscessus_ATCC_199 LGIER----AVLGGTSVGANISIEAAALAPERVAGLLLEGPFLEHGIGAA
MLBr_02603|M.leprae_Br4923 ATRDHPGLKRIVAGHSMGGAIVFAYGVERPDNYDLMVLSGPAVAAQDMVS
* * *. . .
Mb3600c|M.bovis_AF2122/97 PD-PTEG--VKRLSKFSVAPTRENLEAFLR---------VMVYDKNLITP
Rv3569c|M.tuberculosis_H37Rv PD-PTEG--VKRLSKFSVAPTRENLEAFLR---------VMVYDKNLITP
Mvan_4414|M.vanbaalenii_PYR-1 PDGLTEG--LKTLLKGYTDPSPTSMRELVE---------IMTYDSANATD
Mflv_3146|M.gilvum_PYR-GCK DTNPGG--LPKSVFDDFQHQVATNRAGFYRAVPE-----GPFYGFNREGV
TH_3727|M.thermoresistible__bu DANPGG--LPKSVFDDFQAQLAANRSEFYRALAS-----GPFYGFNRDGV
MMAR_1120|M.marinum_M TRSPLGEFLKNPALEAVRFTARSAPKLMHRGRTASRSLIGPILRAASYSD
MUL_0880|M.ulcerans_Agy99 TRSPLGEFLKNPALEAVRFTARSAPKLMHRGRTASRSLIGPILRAASYSD
MAV_4371|M.avium_104 SRSPLGEILKNPALEAVRVAARSAPKLMHRGRNVSRSLIGPVLRAASFSD
MSMEG_3289|M.smegmatis_MC2_155 ISTVTRS--PRQMLNSVLGAIYDMHS-------------DPLHMQIGALN
MAB_1875c|M.abscessus_ATCC_199 GYLWSAGLTLFTLGRPLVWLA------------------GAVARSFPETE
MLBr_02603|M.leprae_Br4923 PLRAVVG-------KGLGLVAPG----------------LPVHQLEVDAI
Mb3600c|M.bovis_AF2122/97 ELVDQRFALASTPESLTATRAMGKSFAGADFEAGMMWREVYRLRQPVLLI
Rv3569c|M.tuberculosis_H37Rv ELVDQRFALASTPESLTATRAMGKSFAGADFEAGMMWREVYRLRQPVLLI
Mvan_4414|M.vanbaalenii_PYR-1 ELVQQRYAAAIGRPDHLENFMIAGLLG----IPQATVEEIAGIETPTLIF
Mflv_3146|M.gilvum_PYR-GCK EPVEGIIANWWRQGMQGGAKAHYDGIVAFSQTDFTED--LQKIRIPVLVM
TH_3727|M.thermoresistible__bu EPSEALIANWWRQGMMGDAKAHYDGIVAFSQTDFTED--LTKITVPVLVM
MMAR_1120|M.marinum_M LEVSRSLDAFSQQMMNDTPIATLVGFLHALEVHDETAGLWTLLRIPTLIA
MUL_0880|M.ulcerans_Agy99 LEVSRSLDAFSQQMMNDTPIATLGGFLHALEVHDETAGLWTLLRIPTLIA
MAV_4371|M.avium_104 LQVSRSLDAFSQRMMNSTPIPTMVGFLDALEHHDETAGLWTLLRVPTLIA
MSMEG_3289|M.smegmatis_MC2_155 RYLRMVAAVTMGKSFASRGFTGAARAIMQSGACAALLAAMREHRIPTIVL
MAB_1875c|M.abscessus_ATCC_199 QPTLGLIRAFLTAPPARSAAFMRGMLVGRMAPPRAVR---RSVNVPTMIL
MLBr_02603|M.leprae_Br4923 SRNRAVVAAYKDDPLVYHGKVPAGVGRVMLQVGETMTRRAPVLTTPLLVV
* ::
Mb3600c|M.bovis_AF2122/97 WGREDRVNPLDGALVALKTIPR--AQLHVFGQCGHWVQVE-KFDEFNKLT
Rv3569c|M.tuberculosis_H37Rv WGREDRVNPLDGALVALKTIPR--AQLHVFGQCGHWVQVE-KFDEFNKLT
Mvan_4414|M.vanbaalenii_PYR-1 HGKDDRVVSYEAALKLVSLIRD--SRVVLINRCGHWLQTE-HSDEFNRLV
Mflv_3146|M.gilvum_PYR-GCK HGDDDQIVPYDDAGPLSAALLPN-GILKTYNGFPHGMPTT-HADVINADL
TH_3727|M.thermoresistible__bu HGDDDQVVPYANSGPLTAELLRN-AKLKTYQGFPHGMMTT-HAEVVNADI
MMAR_1120|M.marinum_M CGDHDLMTPDEYSRKMAASLPQ--SELVIVGGASHLALLD-KPDAINEGL
MUL_0880|M.ulcerans_Agy99 CGDHDLMTPDEYSRKMAASLPQ--SELVIVGGASHLALLD-EPDAINGGL
MAV_4371|M.avium_104 CGDHDLLTPDEYSRKMAASLPQ--SELVIVAGASHLALLD-KPEAINGGL
MSMEG_3289|M.smegmatis_MC2_155 HGEHDSIVPFDNACDVADVVDA---TLYRVRGACHSWLIG-NPRRGADSI
MAB_1875c|M.abscessus_ATCC_199 SLTADPLHPASDAHSLAVDIVG----AQVVSAGTLATLRF-WPRRVTPAI
MLBr_02603|M.leprae_Br4923 HGSEDRLVLVDGSHRLVECVGSTDVELKVYPGLYHEVFNEPERDQVLEDV
* : : :
Mb3600c|M.bovis_AF2122/97 IEFLGGGR------------------------------------------
Rv3569c|M.tuberculosis_H37Rv IEFLGGGR------------------------------------------
Mvan_4414|M.vanbaalenii_PYR-1 ADFVSNR-------------------------------------------
Mflv_3146|M.gilvum_PYR-GCK LEFIRS--------------------------------------------
TH_3727|M.thermoresistible__bu LEFLRS--------------------------------------------
MMAR_1120|M.marinum_M VRLVDRALPGKLTLRYRRVGERLRDRWRRHG-------------------
MUL_0880|M.ulcerans_Agy99 VRLVDRALPGKLTLRYRRVGERLRDRWRRHG-------------------
MAV_4371|M.avium_104 VRLVRRATPSRAALALRRIGERL---WRRG--------------------
MSMEG_3289|M.smegmatis_MC2_155 RQLLHGELGDVLREAATTLGIDDWRDAEDWDRTLVTSAAQVRTLAGAGVV
MAB_1875c|M.abscessus_ATCC_199 VSFLVETFGIDVTARQADA-------------------------------
MLBr_02603|M.leprae_Br4923 VCWILKRL------------------------------------------
:
Mb3600c|M.bovis_AF2122/97 ----------------------------------
Rv3569c|M.tuberculosis_H37Rv ----------------------------------
Mvan_4414|M.vanbaalenii_PYR-1 ----------------------------------
Mflv_3146|M.gilvum_PYR-GCK ----------------------------------
TH_3727|M.thermoresistible__bu ----------------------------------
MMAR_1120|M.marinum_M ----------------------------------
MUL_0880|M.ulcerans_Agy99 ----------------------------------
MAV_4371|M.avium_104 ----------------------------------
MSMEG_3289|M.smegmatis_MC2_155 IGPDSRERVEMDLVRRPGPTRAEAVPETPERATA
MAB_1875c|M.abscessus_ATCC_199 ----------------------------------
MLBr_02603|M.leprae_Br4923 ----------------------------------