For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLQTSQPAPAAESVDRGTSGRPPEVPSWRRKLRPYILSVPAVLIVIGILYPFAVGAYYAFLNYAAVNPD PHFVWFENFKSVLGDQIFWQSVKVTAIFAVLATVIETVIGVGLALLLNRSSLIGKIFEKVLILPLMIAPV IAGVIWKLMFNPQFGILNHVLGLGNTFDWLSSTNALWSVILVDLWIFTPFVAILVLAGIRSLPKEPFEAS EVDGASWFYMFRKLMLPMLWPYILVAVIFRFMDNLKVFDHIYVLTAGGPGVATRTLQIGAFEDSIINLDY SRGSTYMLLLWVIVFITARYLVSVLGKAQRRAAGAES
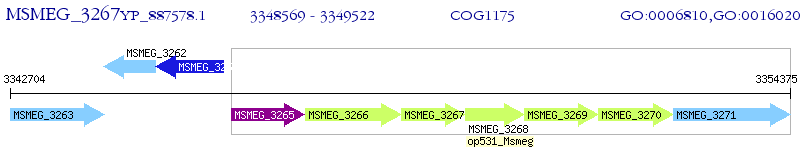
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3267 | - | - | 100% (317) | transporter |
| M. smegmatis MC2 155 | MSMEG_3110 | - | 1e-43 | 33.99% (253) | binding-protein-dependent transport systems inner membrane |
| M. smegmatis MC2 155 | MSMEG_5573 | - | 3e-39 | 31.39% (309) | sugar ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1268 | sugA | 5e-33 | 29.35% (276) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3776 | - | 6e-44 | 31.41% (277) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1236 | sugA | 5e-33 | 29.35% (276) | sugar-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_01087 | - | 6e-33 | 28.47% (274) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1373 | - | 2e-37 | 30.39% (283) | sugar ABC transporter, permease protein SugA |
| M. marinum M | MMAR_4205 | sugA | 6e-34 | 27.82% (284) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1375 | sugA | 2e-31 | 28.73% (275) | ABC transporter, permease protein SugA |
| M. thermoresistible (build 8) | TH_2010 | - | 6e-39 | 32.38% (281) | sugar ABC transporter permease protein |
| M. ulcerans Agy99 | MUL_4512 | sugA | 1e-31 | 29.91% (234) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2623 | - | 2e-42 | 31.13% (257) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1268|M.bovis_AF2122/97 -------------MTSVEQRTATAVFSRTGSRMAERRLAFMLVAPAAMLM
Rv1236|M.tuberculosis_H37Rv -------------MTSVEQRTATAVFSRTGSRMAERRLAFMLVAPAAMLM
MAV_1375|M.avium_104 ------------------------MLGRT----SEQRLALVLVAPAAILM
MLBr_01087|M.leprae_Br4923 -------------MTAVVG---KSWHVRASSVQPEQRLAFLLVTPAAMLM
MMAR_4205|M.marinum_M ----------------------MTALMRSRGRRAERRLAFALVAPATILM
MUL_4512|M.ulcerans_Agy99 ----------------------MTAFLRSRGRRAERRLAFALVAPATILM
MAB_1373|M.abscessus_ATCC_1997 ----------------------MTDRTSSEGKRAERRLGLLLIAPAALLM
Mflv_3776|M.gilvum_PYR-GCK -----------------------------------MKFQHGMVAPLVALF
Mvan_2623|M.vanbaalenii_PYR-1 -------------------------------MMPSLRFQHGMVAPLAALF
TH_2010|M.thermoresistible__bu MTAIAPGASEHLTGSEADRVARIKHAKQQGVSRAEGWRRRGPLLPALIFM
MSMEG_3267|M.smegmatis_MC2_155 -----MTLQTSQPAPAAESVDRGTSGRPPEVPSWRRKLRPYILSVPAVLI
: ::
Mb1268|M.bovis_AF2122/97 VAVTAYPIGYALWLSLQRNNLATPNDTAFIGLGNYHTILIDRYWWTALAV
Rv1236|M.tuberculosis_H37Rv VAVTAYPIGYALWLSLQRNNLATPNDTAFIGLGNYHTILIDRYWWTALAV
MAV_1375|M.avium_104 LAVTAYPIGYAVWLSLQRNNLAAPHDTAFVGLSNYATILSDRYWWTALAV
MLBr_01087|M.leprae_Br4923 LVVTAYPIGYAVWLSLQRYSLATPNETVFIGLGNYQTILTDPYWWTALAV
MMAR_4205|M.marinum_M LLVIGYPIGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAV
MUL_4512|M.ulcerans_Agy99 LLVIGYPIGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAV
MAB_1373|M.abscessus_ATCC_1997 LAVTAYPICYAIWLSVQKYSFASPQDRKFVWFDNYITVLSDRYWWSALVV
Mflv_3776|M.gilvum_PYR-GCK VGVVGFPLGYAAYLSVTDYKLTDQGAPDIVGADNFVATFGDSGFWNAFGT
Mvan_2623|M.vanbaalenii_PYR-1 VGVVGFPLGYAAYLSVTDYKLTDRGAPNLVGADNYVAAFSDGAFWGAFGT
TH_2010|M.thermoresistible__bu IIVTQVPFLFTLYFSTLSWNLVRPGSREFVGFQNYVDVVQDSTFWQVAVN
MSMEG_3267|M.smegmatis_MC2_155 VIGILYPFAVGAYYAFLNYAAVNP-DPHFVWFENFKSVLGDQIFWQSVKV
: *: : : . :: *: . * :*
Mb1268|M.bovis_AF2122/97 TLAITAVSVTIEFVLGLALALVMHRTLIGKGLVRTAVLIPYGIVTVVASY
Rv1236|M.tuberculosis_H37Rv TLAITAVSVTIEFVLGLALALVMHRTLIGKGLVRTAVLIPYGIVTVVASY
MAV_1375|M.avium_104 TLGITVVSVSAEFVLGLALALVMHRTLVGKGLVRTAVLIPYGIVTAVASY
MLBr_01087|M.leprae_Br4923 TLAITVVSVSIEFILGLMLALVMHRTLLGKSLVRIAVLIPYSIVTVVASY
MMAR_4205|M.marinum_M TAAITVVSVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASY
MUL_4512|M.ulcerans_Agy99 TAAITVVSVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASY
MAB_1373|M.abscessus_ATCC_1997 TLVITVISVLIELVLGLALALVMHRTIFGRGVVRTAVLIPYGIVTVAASY
Mflv_3776|M.gilvum_PYR-GCK TALYVLVAVGLELVIGLAIALALQKQRWARDLTRSMLLAPMFITPIAVGL
Mvan_2623|M.vanbaalenii_PYR-1 TALYVLVAVGLELVIGLAIALALQKQRWAKDLTRSMLLAPMFITPIAVGL
TH_2010|M.thermoresistible__bu SVLIIVVTVLVSVLLGLGLALLLDRVFLGRGIVRTLLITPFLVTPVAGAL
MSMEG_3267|M.smegmatis_MC2_155 TAIFAVLATVIETVIGVGLALLLNRSSLIGKIFEKVLILPLMIAPVIAGV
: ::. . ::*: :** :.: : . :: * :.. .
Mb1268|M.bovis_AF2122/97 SWY-YAWTPGTGYLANLLPYDSAPLTQQIP--SLGIVVIAEVWKTTPFMS
Rv1236|M.tuberculosis_H37Rv SWY-YAWTPGTGYLANLLPYDSAPLTQQIP--SLGIVVIAEVWKTTPFMS
MAV_1375|M.avium_104 SWY-YAWTPGTGYLANLLPHGSAPLTAQIP--SLAIVVLAEVWKTTPFMS
MLBr_01087|M.leprae_Br4923 SWY-YAWTPGTGYLASLLPQGSVPLTEQIP--SLSIVIIVEVWKTTPFMS
MMAR_4205|M.marinum_M SWY-YAWTPGTGYLANLLPQGSAPLTQQIP--SLGIVVIAEVWKTTPFMS
MUL_4512|M.ulcerans_Agy99 SWY-YAWTPGTGYLANLLPQGIAPLTQQIP--SLGIVVIAEVWKTTPFMS
MAB_1373|M.abscessus_ATCC_1997 SWY-YAWTPGTGYLANLLPAGSAPLTQQIP--SLAVVILAEVWKTTPFMA
Mflv_3776|M.gilvum_PYR-GCK TFR-FLLNDQLGAIPAMLHAIGIDYDFFGPGRALYTLAFIDVWQWTPFMV
Mvan_2623|M.vanbaalenii_PYR-1 TFR-FLLNDQLGAIPAMLDAIGVEYDFFGPGRALFTLAFIDVWQWTPFMV
TH_2010|M.thermoresistible__bu MWKTAMLDPVFGIVNWLLSPFGVHQVDWVSRFPLTSVMINLVWQWTPFMM
MSMEG_3267|M.smegmatis_MC2_155 IWK-LMFNPQFGILNHVLG-LGNTFDWLSSTNALWSVILVDLWIFTPFVA
: * : :* . .* : : :* ***:
Mb1268|M.bovis_AF2122/97 LLLLAGLALVPEDLLRAAQVDGASAWRRLTKVILPMIKPAIVVALLFRTL
Rv1236|M.tuberculosis_H37Rv LLLLAGLALVPEDLLRAAQVDGASAWRRLTKVILPMIKPAIVVALLFRTL
MAV_1375|M.avium_104 LLLLAGLALVPEDLLKAAQVDGAGAWRRLTRVTLPIIKPAVVVALLFRTL
MLBr_01087|M.leprae_Br4923 LLLLVGLALVPEDLLKAAQMDGAGAWRRLTKIILPIIKPAVMVALLFRTL
MMAR_4205|M.marinum_M LLLLAGLAMVPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFRTL
MUL_4512|M.ulcerans_Agy99 LLLLAGLAMVPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFRTL
MAB_1373|M.abscessus_ATCC_1997 LLLLAGLALVPDDLLKAAQVDGANGWSRLIRVTVPIMKPAILVALLFRTL
Mflv_3776|M.gilvum_PYR-GCK LLLLAGLESIPKEPLDAARVDGARGLYVLRRVTLPLLAPVLVVAILLRSL
Mvan_2623|M.vanbaalenii_PYR-1 LLLLAGLESIPKEPLDAARLDGASGLYVLRRVTLPLLAPVLVVAILLRCL
TH_2010|M.thermoresistible__bu LLILAGLQSMPRDILEAGRVDGAGTFQLFRELTLPHLRRFIELGAVLGAI
MSMEG_3267|M.smegmatis_MC2_155 ILVLAGIRSLPKEPFEASEVDGASWFYMFRKLMLPMLWPYILVAVIFRFM
:*:*.*: :* : : *..:*** : .: :* : : :. :: :
Mb1268|M.bovis_AF2122/97 DAFRIFDNIYVLTGGS--NNTGSVSILGYDNLFKGFNVGLGSA-------
Rv1236|M.tuberculosis_H37Rv DAFRIFDNIYVLTGGS--NNTGSVSILGYDNLFKGFNVGLGSA-------
MAV_1375|M.avium_104 DAFRIFDNIYVLTNGA--NNTGSVSMLGYDNLFKGFNVGLGSA-------
MLBr_01087|M.leprae_Br4923 DAFRIFDNIYVLTRGV--NNTDSVSILGYDNLFKGFNVGLGSA-------
MMAR_4205|M.marinum_M DAFRIFDNIYVLTGGD--NDTASVSILGYDNLFKGFNVGLGSA-------
MUL_4512|M.ulcerans_Agy99 DAFRIFDNIYVLTGGD--NDTASVSILGYDNLFKGFNVGLAVQGIQCGPR
MAB_1373|M.abscessus_ATCC_1997 DAFRIFDNIYVLTGGS--NNTGSVSILGYDNLFKAFNVGLGSA-------
Mflv_3776|M.gilvum_PYR-GCK DAMKVFEYVFATTRGGPGTETETLQYFIYQTGIQFFRLGSASS-------
Mvan_2623|M.vanbaalenii_PYR-1 DAMKVFEYVFATTRGGPGTETETLQYFIYQTGIQFFRLGSASS-------
TH_2010|M.thermoresistible__bu YLVNTFDPIYMMTQGGPGTASSNLPFYIYQRAFLGFDIGQAAA-------
MSMEG_3267|M.smegmatis_MC2_155 DNLKVFDHIYVLTAGGPGVATRTLQIGAFEDSIINLDYSRGST-------
.. *: :: * * : .: :: : : . .
Mb1268|M.bovis_AF2122/97 ---------ISVLIFGCVAVIAFIFIKLFGA----AAPGGEPSGR-----
Rv1236|M.tuberculosis_H37Rv ---------ISVLIFGCVAVIAFIFIKLFGA----AAPGGEPSGR-----
MAV_1375|M.avium_104 ---------ISVLIFGCVGLIALVFVKVFGA----AAPGGDVDGR-----
MLBr_01087|M.leprae_Br4923 ---------ISVLIFGCVALIAVIFIKVFGV----VAPGGDSNGY-----
MMAR_4205|M.marinum_M ---------ISVLIFGCVTLIAVVFITLFGAHQFHSAPGGVTDAR-----
MUL_4512|M.ulcerans_Agy99 FGDQRVDLRLCHSHCGCLHHAVRRPSIRFCARWCDRCALRLGPCRRVAGG
MAB_1373|M.abscessus_ATCC_1997 ---------ISVLIFLCVAIIAVIFIKGFGAS--APTTDGEEARR-----
Mflv_3776|M.gilvum_PYR-GCK ------------MAFVVLIVVLAAIVFAFRR----MDRAREAGIR-----
Mvan_2623|M.vanbaalenii_PYR-1 ------------MAFIVLVVVLAVIVIAFRR----MERTKSR--------
TH_2010|M.thermoresistible__bu --------------MGVIVVIFTIIIATFALRLIFRSFTGKEEAA-----
MSMEG_3267|M.smegmatis_MC2_155 --------------YMLLLWVIVFITARYLVSVLGKAQRRAAGAES----
: :
Mb1268|M.bovis_AF2122/97 ----------------------
Rv1236|M.tuberculosis_H37Rv ----------------------
MAV_1375|M.avium_104 ----------------------
MLBr_01087|M.leprae_Br4923 ----------------------
MMAR_4205|M.marinum_M ----------------------
MUL_4512|M.ulcerans_Agy99 ERSGWPSTPWSWSTPCSRCCGS
MAB_1373|M.abscessus_ATCC_1997 ----------------------
Mflv_3776|M.gilvum_PYR-GCK ----------------------
Mvan_2623|M.vanbaalenii_PYR-1 ----------------------
TH_2010|M.thermoresistible__bu ----------------------
MSMEG_3267|M.smegmatis_MC2_155 ----------------------