For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKFQHGMVAPLVALFVGVVGFPLGYAAYLSVTDYKLTDQGAPDIVGADNFVATFGDSGFWNAFGTTALYV LVAVGLELVIGLAIALALQKQRWARDLTRSMLLAPMFITPIAVGLTFRFLLNDQLGAIPAMLHAIGIDYD FFGPGRALYTLAFIDVWQWTPFMVLLLLAGLESIPKEPLDAARVDGARGLYVLRRVTLPLLAPVLVVAIL LRSLDAMKVFEYVFATTRGGPGTETETLQYFIYQTGIQFFRLGSASSMAFVVLIVVLAAIVFAFRRMDRA REAGIR
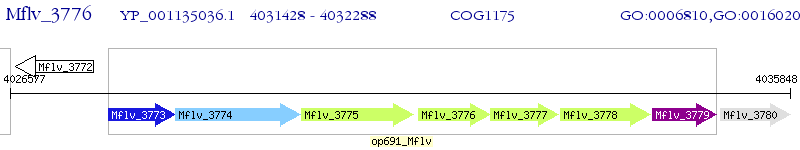
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3776 | - | - | 100% (286) | binding-protein-dependent transport systems inner membrane component |
| M. gilvum PYR-GCK | Mflv_3852 | - | 1e-36 | 30.60% (281) | binding-protein-dependent transport systems inner membrane |
| M. gilvum PYR-GCK | Mflv_3777 | - | 6e-10 | 25.89% (282) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1268 | sugA | 1e-33 | 30.94% (278) | sugar-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2835c | ugpA | 2e-34 | 33.45% (278) | sn-glycerol-3-phosphate transport integral membrane protein |
| M. leprae Br4923 | MLBr_01087 | - | 3e-34 | 28.84% (267) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1373 | - | 1e-37 | 32.58% (267) | sugar ABC transporter, permease protein SugA |
| M. marinum M | MMAR_4205 | sugA | 2e-35 | 31.84% (267) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1375 | sugA | 1e-34 | 32.96% (267) | ABC transporter, permease protein SugA |
| M. smegmatis MC2 155 | MSMEG_3110 | - | 1e-116 | 70.21% (282) | binding-protein-dependent transport systems inner membrane |
| M. thermoresistible (build 8) | TH_2010 | - | 4e-41 | 31.29% (278) | sugar ABC transporter permease protein |
| M. ulcerans Agy99 | MUL_4512 | sugA | 3e-33 | 32.13% (249) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2623 | - | 1e-145 | 90.39% (281) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1268|M.bovis_AF2122/97 -------------MTSVEQRTATAVFSRTGSRMAERRLAFMLVAPAAMLM
MAV_1375|M.avium_104 ------------------------MLGRT----SEQRLALVLVAPAAILM
MLBr_01087|M.leprae_Br4923 -------------MTAVVG---KSWHVRASSVQPEQRLAFLLVTPAAMLM
MMAR_4205|M.marinum_M ----------------------MTALMRSRGRRAERRLAFALVAPATILM
MUL_4512|M.ulcerans_Agy99 ----------------------MTAFLRSRGRRAERRLAFALVAPATILM
MAB_1373|M.abscessus_ATCC_1997 ----------------------MTDRTSSEGKRAERRLGLLLIAPAALLM
Mflv_3776|M.gilvum_PYR-GCK --------------------MKFQHG---------------MVAPLVALF
Mvan_2623|M.vanbaalenii_PYR-1 ----------------MMPSLRFQHG---------------MVAPLAALF
MSMEG_3110|M.smegmatis_MC2_155 --------------------MKFKYG---------------MLMPLLALF
Rv2835c|M.tuberculosis_H37Rv --------------MAAPQRARLRSSKERVRDYAL---FVVLVGPNVALL
TH_2010|M.thermoresistible__bu MTAIAPGASEHLTGSEADRVARIKHAKQQGVSRAEGWRRRGPLLPALIFM
: * ::
Mb1268|M.bovis_AF2122/97 VAVTAYPIGYALWLSLQRNNLATPNDTAFIGLGNYHTILIDRYWWTALAV
MAV_1375|M.avium_104 LAVTAYPIGYAVWLSLQRNNLAAPHDTAFVGLSNYATILSDRYWWTALAV
MLBr_01087|M.leprae_Br4923 LVVTAYPIGYAVWLSLQRYSLATPNETVFIGLGNYQTILTDPYWWTALAV
MMAR_4205|M.marinum_M LLVIGYPIGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAV
MUL_4512|M.ulcerans_Agy99 LLVIGYPIGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAV
MAB_1373|M.abscessus_ATCC_1997 LAVTAYPICYAIWLSVQKYSFASPQDRKFVWFDNYITVLSDRYWWSALVV
Mflv_3776|M.gilvum_PYR-GCK VGVVGFPLGYAAYLSVTDYKLTDQGAPDIVGADNFVATFGDSGFWNAFGT
Mvan_2623|M.vanbaalenii_PYR-1 VGVVGFPLGYAAYLSVTDYKLTDRGAPNLVGADNYVAAFSDGAFWGAFGT
MSMEG_3110|M.smegmatis_MC2_155 AVAVGFPLCYAAYLSVTDYKLTDREPPDFVGTANFATALSNPAFWEAFAT
Rv2835c|M.tuberculosis_H37Rv LLFVYRPLADNIRLSFFDWNVSDPSAR-FVGLSNYTEWFTRSDTRQIVFN
TH_2010|M.thermoresistible__bu IIVTQVPFLFTLYFSTLSWNLVRPGSREFVGFQNYVDVVQDSTFWQVAVN
*: :* .. :: *: .
Mb1268|M.bovis_AF2122/97 TLAITAVSVTIEFVLGLALALVMHRTLIGKGLVRTAVLIPYGIVTVVASY
MAV_1375|M.avium_104 TLGITVVSVSAEFVLGLALALVMHRTLVGKGLVRTAVLIPYGIVTAVASY
MLBr_01087|M.leprae_Br4923 TLAITVVSVSIEFILGLMLALVMHRTLLGKSLVRIAVLIPYSIVTVVASY
MMAR_4205|M.marinum_M TAAITVVSVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASY
MUL_4512|M.ulcerans_Agy99 TAAITVVSVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASY
MAB_1373|M.abscessus_ATCC_1997 TLVITVISVLIELVLGLALALVMHRTIFGRGVVRTAVLIPYGIVTVAASY
Mflv_3776|M.gilvum_PYR-GCK TALYVLVAVGLELVIGLAIALALQKQRWARDLTRSMLLAPMFITPIAVGL
Mvan_2623|M.vanbaalenii_PYR-1 TALYVLVAVGLELVIGLAIALALQKQRWAKDLTRSMLLAPMFITPIAVGL
MSMEG_3110|M.smegmatis_MC2_155 TATYVVIAVCGELVIGFALALALHQQRWAKDLSRAIVLAPMFITPIAVGL
Rv2835c|M.tuberculosis_H37Rv TAVFTGAAVVGSMVLGLALAMLLDRPLRGRNLVRSTVFAPFVISGAAVGL
TH_2010|M.thermoresistible__bu SVLIIVVTVLVSVLLGLGLALLLDRVFLGRGIVRTLLITPFLVTPVAGAL
: :* ..::*: :*: :.: .:.: * :: * : . .
Mb1268|M.bovis_AF2122/97 SWY-YAWTPGTGYLANLLPYDSAPLTQQIPSLG----IVVIAEVWKTTPF
MAV_1375|M.avium_104 SWY-YAWTPGTGYLANLLPHGSAPLTAQIPSLA----IVVLAEVWKTTPF
MLBr_01087|M.leprae_Br4923 SWY-YAWTPGTGYLASLLPQGSVPLTEQIPSLS----IVIIVEVWKTTPF
MMAR_4205|M.marinum_M SWY-YAWTPGTGYLANLLPQGSAPLTQQIPSLG----IVVIAEVWKTTPF
MUL_4512|M.ulcerans_Agy99 SWY-YAWTPGTGYLANLLPQGIAPLTQQIPSLG----IVVIAEVWKTTPF
MAB_1373|M.abscessus_ATCC_1997 SWY-YAWTPGTGYLANLLPAGSAPLTQQIPSLA----VVILAEVWKTTPF
Mflv_3776|M.gilvum_PYR-GCK TFR-FLLNDQLGAIPAMLHAIGIDY-DFFGPGR-ALYTLAFIDVWQWTPF
Mvan_2623|M.vanbaalenii_PYR-1 TFR-FLLNDQLGAIPAMLDAIGVEY-DFFGPGR-ALFTLAFIDVWQWTPF
MSMEG_3110|M.smegmatis_MC2_155 IFR-FLLNDQIGAVPALLHAVGADH-DFFGSGR-ALLTLAFIDIWQWTPF
Rv2835c|M.tuberculosis_H37Rv AAQ-FVFDPHFGLIQDLLRRIGVGVPDFYQDARWALFMVTITYVWKNLGY
TH_2010|M.thermoresistible__bu MWKTAMLDPVFGIVNWLLSPFGVHQVDWVSRFP--LTSVMINLVWQWTPF
* : :* : : :*: :
Mb1268|M.bovis_AF2122/97 MSLLLLAGLALVPEDLLRAAQVDGASAWRRLTKVILPMIKPAIVVALLFR
MAV_1375|M.avium_104 MSLLLLAGLALVPEDLLKAAQVDGAGAWRRLTRVTLPIIKPAVVVALLFR
MLBr_01087|M.leprae_Br4923 MSLLLLVGLALVPEDLLKAAQMDGAGAWRRLTKIILPIIKPAVMVALLFR
MMAR_4205|M.marinum_M MSLLLLAGLAMVPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFR
MUL_4512|M.ulcerans_Agy99 MSLLLLAGLAMVPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFR
MAB_1373|M.abscessus_ATCC_1997 MALLLLAGLALVPDDLLKAAQVDGANGWSRLIRVTVPIMKPAILVALLFR
Mflv_3776|M.gilvum_PYR-GCK MVLLLLAGLESIPKEPLDAARVDGARGLYVLRRVTLPLLAPVLVVAILLR
Mvan_2623|M.vanbaalenii_PYR-1 MVLLLLAGLESIPKEPLDAARLDGASGLYVLRRVTLPLLAPVLVVAILLR
MSMEG_3110|M.smegmatis_MC2_155 MVLLILAGLESMPKQPMEAARVDGAGPVYRLRRVLIPMLAPVLTVAVLLR
Rv2835c|M.tuberculosis_H37Rv TFVIYLAALQGVRRDLLEAAEIDGASRWAVFRRVLLPQLRPTTFFLSITV
TH_2010|M.thermoresistible__bu MMLLILAGLQSMPRDILEAGRVDGAGTFQLFRELTLPHLRRFIELGAVLG
:: *..* : : : *..:*** : .: :* : . :
Mb1268|M.bovis_AF2122/97 TLDAFRIFDNIYVLTGGSN---NTGSVSILGYDNLFKGFNVGLGSA----
MAV_1375|M.avium_104 TLDAFRIFDNIYVLTNGAN---NTGSVSMLGYDNLFKGFNVGLGSA----
MLBr_01087|M.leprae_Br4923 TLDAFRIFDNIYVLTRGVN---NTDSVSILGYDNLFKGFNVGLGSA----
MMAR_4205|M.marinum_M TLDAFRIFDNIYVLTGGDN---DTASVSILGYDNLFKGFNVGLGSA----
MUL_4512|M.ulcerans_Agy99 TLDAFRIFDNIYVLTGGDN---DTASVSILGYDNLFKGFNVGLAVQGIQC
MAB_1373|M.abscessus_ATCC_1997 TLDAFRIFDNIYVLTGGSN---NTGSVSILGYDNLFKAFNVGLGSA----
Mflv_3776|M.gilvum_PYR-GCK SLDAMKVFEYVFATTRGGP-GTETETLQYFIYQTGIQFFRLGSASS----
Mvan_2623|M.vanbaalenii_PYR-1 CLDAMKVFEYVFATTRGGP-GTETETLQYFIYQTGIQFFRLGSASS----
MSMEG_3110|M.smegmatis_MC2_155 GLDALRVFEYVYATTRGGP-GTETQTLQYYMYLEGIQFFRLAQASA----
Rv2835c|M.tuberculosis_H37Rv LINSLQVFDVINVMTRGGPEGTGTTTMVYQVYVETFRNFRAGYGAT----
TH_2010|M.thermoresistible__bu AIYLVNTFDPIYMMTQGGP-GTASSNLPFYIYQRAFLGFDIGQAAA----
: .. *: : * * : .: * : * . .
Mb1268|M.bovis_AF2122/97 ------------ISVLIFGCVAVIAFIFIKLFGA----AAPGGEPSGR--
MAV_1375|M.avium_104 ------------ISVLIFGCVGLIALVFVKVFGA----AAPGGDVDGR--
MLBr_01087|M.leprae_Br4923 ------------ISVLIFGCVALIAVIFIKVFGV----VAPGGDSNGY--
MMAR_4205|M.marinum_M ------------ISVLIFGCVTLIAVVFITLFGAHQFHSAPGGVTDAR--
MUL_4512|M.ulcerans_Agy99 GPRFGDQRVDLRLCHSHCGCLHHAVRRPSIRFCARWCDRCALRLGPCRRV
MAB_1373|M.abscessus_ATCC_1997 ------------ISVLIFLCVAIIAVIFIKGFGAS--APTTDGEEARR--
Mflv_3776|M.gilvum_PYR-GCK -----------------MAFVVLIVVLAAIVFAFRRMDRAREAGIR----
Mvan_2623|M.vanbaalenii_PYR-1 -----------------MAFIVLVVVLAVIVIAFRRMERTKSR-------
MSMEG_3110|M.smegmatis_MC2_155 -----------------MAFIVLVLVLAVIVVAFRTMERTRERTPSR---
Rv2835c|M.tuberculosis_H37Rv -----------------VATIMFLVLLAVTYYQVRVMDRGQRQ-------
TH_2010|M.thermoresistible__bu -----------------MGVIVVIFTIIIATFALRLIFRSFTGKEEAA--
:
Mb1268|M.bovis_AF2122/97 -------------------------
MAV_1375|M.avium_104 -------------------------
MLBr_01087|M.leprae_Br4923 -------------------------
MMAR_4205|M.marinum_M -------------------------
MUL_4512|M.ulcerans_Agy99 AGGERSGWPSTPWSWSTPCSRCCGS
MAB_1373|M.abscessus_ATCC_1997 -------------------------
Mflv_3776|M.gilvum_PYR-GCK -------------------------
Mvan_2623|M.vanbaalenii_PYR-1 -------------------------
MSMEG_3110|M.smegmatis_MC2_155 -------------------------
Rv2835c|M.tuberculosis_H37Rv -------------------------
TH_2010|M.thermoresistible__bu -------------------------