For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TALMRSRGRRAERRLAFALVAPATILMLLVIGYPIGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRY WWTALAVTAAITVVSVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASYSWYYAWTPGTGYL ANLLPQGSAPLTQQIPSLGIVVIAEVWKTTPFMSLLLLAGLAMVPEDLLQAAQVDGAGAWRRLTKIILPL IKPAVLVALLFRTLDAFRIFDNIYVLTGGDNDTASVSILGYDNLFKGFNVGLGSAISVLIFGCVTLIAVV FITLFGAHQFHSAPGGVTDAR*
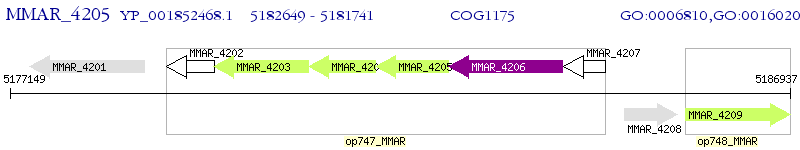
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4205 | sugA | - | 100% (302) | sugar-transport integral membrane protein ABC transporter SugA |
| M. marinum M | MMAR_3617 | uspA | 2e-37 | 33.45% (281) | sugar-transport integral membrane protein ABC transporter |
| M. marinum M | MMAR_3013 | - | 8e-29 | 27.67% (300) | ABC-type sugar transport integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1268 | sugA | 1e-140 | 83.16% (297) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3852 | - | 4e-72 | 49.82% (281) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1236 | sugA | 1e-140 | 83.16% (297) | sugar-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_01087 | - | 1e-132 | 79.86% (293) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1373 | - | 1e-121 | 72.22% (288) | sugar ABC transporter, permease protein SugA |
| M. avium 104 | MAV_1375 | sugA | 1e-135 | 81.29% (294) | ABC transporter, permease protein SugA |
| M. smegmatis MC2 155 | MSMEG_5060 | - | 1e-128 | 75.34% (296) | ABC transporter, permease protein SugA |
| M. thermoresistible (build 8) | TH_4307 | - | 1e-125 | 73.81% (294) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4512 | sugA | 1e-146 | 98.86% (263) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2543 | - | 2e-71 | 49.82% (281) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4205|M.marinum_M ---------------MTALMRSRGRRAERRLAFALVAPATILMLLVIGYP
MUL_4512|M.ulcerans_Agy99 ---------------MTAFLRSRGRRAERRLAFALVAPATILMLLVIGYP
Mb1268|M.bovis_AF2122/97 MTSVEQRTA------TAVFSRTGSRMAERRLAFMLVAPAAMLMVAVTAYP
Rv1236|M.tuberculosis_H37Rv MTSVEQRTA------TAVFSRTGSRMAERRLAFMLVAPAAMLMVAVTAYP
MAV_1375|M.avium_104 -----------------MLGRT----SEQRLALVLVAPAAILMLAVTAYP
MLBr_01087|M.leprae_Br4923 ---MTAVVG------KSWHVRASSVQPEQRLAFLLVTPAAMLMLVVTAYP
MSMEG_5060|M.smegmatis_MC2_155 ---MTAAVT-----PSASAVASDDKKSERRLAFWLIAPAVLLMLAVTAYP
TH_4307|M.thermoresistible__bu ---MTAAAEKADPTPAAAVARTDDTRSERRLAYWLIAPAVLLMVAVTGYP
MAB_1373|M.abscessus_ATCC_1997 ---MTDRTS------------SEGKRAERRLGLLLIAPAALLMLAVTAYP
Mflv_3852|M.gilvum_PYR-GCK ---MTAPADAPIQKDPGKPSVSERAKGERRLGLYLTLPSYVVMLLVTAYP
Mvan_2543|M.vanbaalenii_PYR-1 ---MTALHEAPIQKDPGTPAVSERARGERRLGIYLTLPSYVVMLLVTAYP
: *:**. * *: ::*: * .**
MMAR_4205|M.marinum_M IGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAVTAAITVV
MUL_4512|M.ulcerans_Agy99 IGYAMWLSLQRNNLATPNETRFIGLGNYQTILVDRYWWTALAVTAAITVV
Mb1268|M.bovis_AF2122/97 IGYALWLSLQRNNLATPNDTAFIGLGNYHTILIDRYWWTALAVTLAITAV
Rv1236|M.tuberculosis_H37Rv IGYALWLSLQRNNLATPNDTAFIGLGNYHTILIDRYWWTALAVTLAITAV
MAV_1375|M.avium_104 IGYAVWLSLQRNNLAAPHDTAFVGLSNYATILSDRYWWTALAVTLGITVV
MLBr_01087|M.leprae_Br4923 IGYAVWLSLQRYSLATPNETVFIGLGNYQTILTDPYWWTALAVTLAITVV
MSMEG_5060|M.smegmatis_MC2_155 IGYAVWLSLQRYNLAEPHDTEFIGLANYVTVLTDGYWWTAFAVTLGITVV
TH_4307|M.thermoresistible__bu IAYAVWLSLLRYNLAQPDDVSFVGLENYVTVLTDEYWWTAFAVTLGITVV
MAB_1373|M.abscessus_ATCC_1997 ICYAIWLSVQKYSFASPQDRKFVWFDNYITVLSDRYWWSALVVTLVITVI
Mflv_3852|M.gilvum_PYR-GCK LGYALVLSLYNFRLTDPEGRSFVGLGNYLVILTDPIWWNDFVTTLAITVV
Mvan_2543|M.vanbaalenii_PYR-1 LGYALVLSLYNFRLTDPEGRSFVGLSNYLVILTDPLWWNDFVTTLAITVV
: **: **: . :: *. *: : ** .:* * **. :..* **.:
MMAR_4205|M.marinum_M SVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASYSWYYAWT
MUL_4512|M.ulcerans_Agy99 SVACEFVLGLALALVMHRSLVAKGLVRTAVLVPYGIVTVVASYSWYYAWT
Mb1268|M.bovis_AF2122/97 SVTIEFVLGLALALVMHRTLIGKGLVRTAVLIPYGIVTVVASYSWYYAWT
Rv1236|M.tuberculosis_H37Rv SVTIEFVLGLALALVMHRTLIGKGLVRTAVLIPYGIVTVVASYSWYYAWT
MAV_1375|M.avium_104 SVSAEFVLGLALALVMHRTLVGKGLVRTAVLIPYGIVTAVASYSWYYAWT
MLBr_01087|M.leprae_Br4923 SVSIEFILGLMLALVMHRTLLGKSLVRIAVLIPYSIVTVVASYSWYYAWT
MSMEG_5060|M.smegmatis_MC2_155 SVAIEFALGLALALVMHRTIFGKGAVRTAILIPYGIVTVAASYSWYYAWT
TH_4307|M.thermoresistible__bu SVAIEFVLGLALALVMHRTIFGKGVVRTAILIPYGIVTVAASYSWYYAWT
MAB_1373|M.abscessus_ATCC_1997 SVLIELVLGLALALVMHRTIFGRGVVRTAVLIPYGIVTVAASYSWYYAWT
Mflv_3852|M.gilvum_PYR-GCK TVAIELVLGFWFAFVMLRIVRGRGPLRTAILIPYGIVTVVSAFIWRYAFA
Mvan_2543|M.vanbaalenii_PYR-1 TVAIELVLGFWFAFVMLRIVRGRGPLRTAILIPYGIVTVVSAFIWRYAFA
:* *: **: :*:** * : .:. :* *:*:**.***..::: * **::
MMAR_4205|M.marinum_M PGTGYLANLLPQGSAPLT-QQIPSLGIVVIAEVWKTTPFMSLLLLAGLAM
MUL_4512|M.ulcerans_Agy99 PGTGYLANLLPQGIAPLT-QQIPSLGIVVIAEVWKTTPFMSLLLLAGLAM
Mb1268|M.bovis_AF2122/97 PGTGYLANLLPYDSAPLT-QQIPSLGIVVIAEVWKTTPFMSLLLLAGLAL
Rv1236|M.tuberculosis_H37Rv PGTGYLANLLPYDSAPLT-QQIPSLGIVVIAEVWKTTPFMSLLLLAGLAL
MAV_1375|M.avium_104 PGTGYLANLLPHGSAPLT-AQIPSLAIVVLAEVWKTTPFMSLLLLAGLAL
MLBr_01087|M.leprae_Br4923 PGTGYLASLLPQGSVPLT-EQIPSLSIVIIVEVWKTTPFMSLLLLVGLAL
MSMEG_5060|M.smegmatis_MC2_155 PGTGYLANLLPEGSAPLT-DQLPSLAIVVLAEVWKTTPFMALLLLAGLAL
TH_4307|M.thermoresistible__bu PGTGYLANLLPDGSAPLT-QQLPSLAIVVLAEVWKTTPFMALLLLAGLAL
MAB_1373|M.abscessus_ATCC_1997 PGTGYLANLLPAGSAPLT-QQIPSLAVVILAEVWKTTPFMALLLLAGLAL
Mflv_3852|M.gilvum_PYR-GCK IDSGFVNQWLNLTDFDWFGDRWSAIFVICLSEIWKTTPFISLLLLAGLVQ
Mvan_2543|M.vanbaalenii_PYR-1 IDSGFVNQWLNLTEFDWFGDRWSAIFVICLSEIWKTTPFISLLLLAGLVQ
.:*:: . * : .:: :: : *:******::****.**.
MMAR_4205|M.marinum_M VPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFRTLDAFRIFDNI
MUL_4512|M.ulcerans_Agy99 VPEDLLQAAQVDGAGAWRRLTKIILPLIKPAVLVALLFRTLDAFRIFDNI
Mb1268|M.bovis_AF2122/97 VPEDLLRAAQVDGASAWRRLTKVILPMIKPAIVVALLFRTLDAFRIFDNI
Rv1236|M.tuberculosis_H37Rv VPEDLLRAAQVDGASAWRRLTKVILPMIKPAIVVALLFRTLDAFRIFDNI
MAV_1375|M.avium_104 VPEDLLKAAQVDGAGAWRRLTRVTLPIIKPAVVVALLFRTLDAFRIFDNI
MLBr_01087|M.leprae_Br4923 VPEDLLKAAQMDGAGAWRRLTKIILPIIKPAVMVALLFRTLDAFRIFDNI
MSMEG_5060|M.smegmatis_MC2_155 VPQDLLNAAQVDGAGPWKRLTKVILPMIKPAILVALLFRTLDAFRIFDNI
TH_4307|M.thermoresistible__bu VPQELLNAAQMDGANAWQRLVKIILPMIKPAILVALLFRTLDAFRIFDNI
MAB_1373|M.abscessus_ATCC_1997 VPDDLLKAAQVDGANGWSRLIRVTVPIMKPAILVALLFRTLDAFRIFDNI
Mflv_3852|M.gilvum_PYR-GCK VPEDMQEAAKVDGATAWQRLWKITLPNMKAAIMVALLFRTLDAWRIFDNP
Mvan_2543|M.vanbaalenii_PYR-1 VPEDMQEAAKVDGATAWQRLWKITLPNMKAAIMVALLFRTLDAWRIFDNP
**::: .**::*** * ** :: :* :*.*::**********:*****
MMAR_4205|M.marinum_M YVLTGGDNDTASVSILGYDNLFKGFNVGLGSA----------------IS
MUL_4512|M.ulcerans_Agy99 YVLTGGDNDTASVSILGYDNLFKGFNVGLAVQGIQCGPRFGDQRVDLRLC
Mb1268|M.bovis_AF2122/97 YVLTGGSNNTGSVSILGYDNLFKGFNVGLGSA----------------IS
Rv1236|M.tuberculosis_H37Rv YVLTGGSNNTGSVSILGYDNLFKGFNVGLGSA----------------IS
MAV_1375|M.avium_104 YVLTNGANNTGSVSMLGYDNLFKGFNVGLGSA----------------IS
MLBr_01087|M.leprae_Br4923 YVLTRGVNNTDSVSILGYDNLFKGFNVGLGSA----------------IS
MSMEG_5060|M.smegmatis_MC2_155 YILTGGSNDTGSVSILGYDNLFKAFNVGLGSA----------------IS
TH_4307|M.thermoresistible__bu YILTGGANNTGSVSILGYNNLFKAFNLGLGSA----------------IS
MAB_1373|M.abscessus_ATCC_1997 YVLTGGSNNTGSVSILGYDNLFKAFNVGLGSA----------------IS
Mflv_3852|M.gilvum_PYR-GCK YVMTAGANNTETISFLAYRQNVTLVNLGMGSA----------------VS
Mvan_2543|M.vanbaalenii_PYR-1 YVMTAGANNTETISFLAYRQNVTLVNLGMGSA----------------VS
*::* * *:* ::*:*.* : .. .*:*:. :.
MMAR_4205|M.marinum_M VLIFGCVTLIAVVFITLFGAHQFHSAPGGVTDAR----------------
MUL_4512|M.ulcerans_Agy99 HSHCGCLHHAVRRPSIRFCARWCDRCALRLGPCRRVAGGERSGWPSTPWS
Mb1268|M.bovis_AF2122/97 VLIFGCVAVIAFIFIKLFGA----AAPGGEPSGR----------------
Rv1236|M.tuberculosis_H37Rv VLIFGCVAVIAFIFIKLFGA----AAPGGEPSGR----------------
MAV_1375|M.avium_104 VLIFGCVGLIALVFVKVFGA----AAPGGDVDGR----------------
MLBr_01087|M.leprae_Br4923 VLIFGCVALIAVIFIKVFGV----VAPGGDSNGY----------------
MSMEG_5060|M.smegmatis_MC2_155 VLIFLSVAIIAFIYIKIFGA----AAPGSDEEVR----------------
TH_4307|M.thermoresistible__bu VLIFLCVAVIAFIYIKIFGT----SAPGSDAERS----------------
MAB_1373|M.abscessus_ATCC_1997 VLIFLCVAIIAVIFIKGFGA----SAPTTDGEEARR--------------
Mflv_3852|M.gilvum_PYR-GCK VLLFLSVVAIAWIFIKVFKT----DLSQVRGDQ-----------------
Mvan_2543|M.vanbaalenii_PYR-1 VLLFLSVVGIAWIFIKVFKT----DLSQVRGDQ-----------------
.: . * . .
MMAR_4205|M.marinum_M -----------
MUL_4512|M.ulcerans_Agy99 WSTPCSRCCGS
Mb1268|M.bovis_AF2122/97 -----------
Rv1236|M.tuberculosis_H37Rv -----------
MAV_1375|M.avium_104 -----------
MLBr_01087|M.leprae_Br4923 -----------
MSMEG_5060|M.smegmatis_MC2_155 -----------
TH_4307|M.thermoresistible__bu -----------
MAB_1373|M.abscessus_ATCC_1997 -----------
Mflv_3852|M.gilvum_PYR-GCK -----------
Mvan_2543|M.vanbaalenii_PYR-1 -----------