For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLASGTGRIARFDEPGKPFEIQTVPVPQVGPGEILIRVTRANICGSDVHAWHGTFATRGLGGQLPTVLGH EMVGVVAVLGDDVHSDSDGRPLEVGTRVVFPYFFCCHTCRNCLAGRRNACVNLKMAMLGRADEPPHFVGG YADYFLLPAKAVVYTVPDSVSDEIAAGANCALSQVMYGLERVDLQLGETVVVQGAGALGLYAVAVAKARG AKTVIAIDGVPERLELAKAFGADEVIDITATTDKDRVKIVRRLTDRQGADVVVEVVGHPSAIDEGLKLLG QFGRYVEIGNINIGKTFEFDPSRFVFANKTMVGVSLYDPAVLSRALTFLAEYQDRLPFDRLAAACYRLED INDAFSAAENKRDVRASIVP*
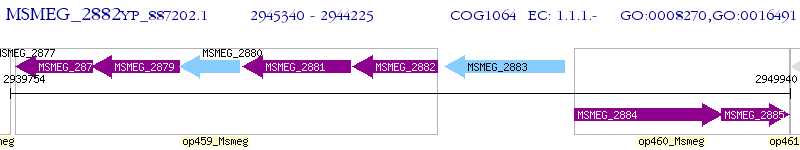
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2882 | - | - | 100% (371) | 5-exo-alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6296 | - | 3e-58 | 36.77% (359) | 5-exo-alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4400 | - | 3e-45 | 37.46% (323) | alcohol dehydrogenase, zinc-containing |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1557 | adh | 1e-31 | 31.32% (348) | alcohol dehydrogenase adh |
| M. gilvum PYR-GCK | Mflv_2766 | - | 4e-30 | 30.39% (362) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1530 | adh | 1e-31 | 31.32% (348) | alcohol dehydrogenase adh |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 5e-26 | 26.78% (366) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_1869c | - | 3e-28 | 28.93% (363) | zinc-dependent alcohol dehydrogenase AdhE2 |
| M. marinum M | MMAR_0405 | adhE | 2e-28 | 28.65% (370) | zinc-type alcohol dehydrogenase (E subunit) AdhE |
| M. avium 104 | MAV_5022 | - | 2e-27 | 29.81% (369) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_4349 | - | 9e-31 | 34.38% (256) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_4777 | - | 2e-26 | 31.94% (288) | zinc-dependent alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2879 | - | 0.0 | 88.49% (365) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2766|M.gilvum_PYR-GCK -------------------MTQTVRGVIARAKGRPVEVVDVVIPDPGPGE
MLBr_01784|M.leprae_Br4923 -------------------MSQTVRGVISRKKDEPVELVDIVIPDPGPGE
MAB_1869c|M.abscessus_ATCC_199 -----------------------------------MELVDIVIPDPGPGE
MMAR_0405|M.marinum_M MELGLQPWLYSNMIHIRGAVLERIGSPRPYAQSRPISVVDLDLDPPGDDE
MAV_5022|M.avium_104 -------------------MLQRIGAPRPYARSRPISITDVELAPPGRDE
MUL_4777|M.ulcerans_Agy99 -------------------MAATMRAERFYADSKTVVLEDVPIPEPGPGE
MSMEG_2882|M.smegmatis_MC2_155 ----------------MTLASGTGRIARFDEPGKPFEIQTVPVPQVGPGE
Mvan_2879|M.vanbaalenii_PYR-1 ----------------MSRAPFTGRIARFDEPGKPFDIETVTLPEVGPGE
Mb1557|M.bovis_AF2122/97 ------------------MSDGAVVRALVLEAPRRLVVRQYRLPRIGDDD
Rv1530|M.tuberculosis_H37Rv ------------------MSDGAVVRALVLEAPRRLVVRQYRLPRIGDDD
TH_4349|M.thermoresistible__bu -----------------------MKALRLMDWKSEPELVEVPDPTPGPGQ
: * .:
Mflv_2766|M.gilvum_PYR-GCK VVVDIQACGVCHTDLTYRDG-----GINDEFPFLLG-HEAAGIVETVGQG
MLBr_01784|M.leprae_Br4923 AVVDVTACGVCHTDLTYREG-----GINDRYPFLLG-HEAAGTVEVVGPG
MAB_1869c|M.abscessus_ATCC_199 VVVKVQACGVCHTDLTYREG-----GINDEFPFLLG-HEAAGIVETVGEG
MMAR_0405|M.marinum_M VLVRIEAAGVCHSDLSVVDG-----NRVRPVPMLLG-HEAAGIVERVGDR
MAV_5022|M.avium_104 VLVRIEAAGICHSDLSVVDG-----NRVRPVPMLLG-HEAAGIVERVGPG
MUL_4777|M.ulcerans_Agy99 VLVKVAYCGICHSDLSLING-----TFPTQAPVVTQGHEASGTIVKLGPE
MSMEG_2882|M.smegmatis_MC2_155 ILIRVTRANICGSDVHAWHGTFATRGLGGQLPTVLG-HEMVGVVAVLGDD
Mvan_2879|M.vanbaalenii_PYR-1 ILIKVTRANICGSDVHAWHGTFATRGLGGQLPTVLG-HEMVGAVEELGAG
Mb1557|M.bovis_AF2122/97 ALVRVEACGLCGTDHEQYTG-----ELAGGFAFVPG-HETVGTIAAIGPR
Rv1530|M.tuberculosis_H37Rv ALVRVEACGLCGTDHEQYTG-----ELAGGFAFVPG-HETVGTIAAIGPR
TH_4349|M.thermoresistible__bu VLVKIGGAGACHSDLHLMHDFEPG-MMPWQLPFTLG-HENAGWVAAVGAG
:: : .. * :* . . ** * : :*
Mflv_2766|M.gilvum_PYR-GCK VT------NVAPGDFVILNWRAVCGQCRACKRGRPHLCFDTHNAAQKMTL
MLBr_01784|M.leprae_Br4923 VT------AVEPGDFVILNWRAVCGQCRACKRGRPSYCFDTFNAQQKMTL
MAB_1869c|M.abscessus_ATCC_199 VT------TVEPGDFVILNWRAVCGVCRACKRGRPQYCFDTFNAAQKMTL
MMAR_0405|M.marinum_M VD------DLAVGQRVVLVFLPRCGHCPSCATEGITPCAPGSATNAAGTL
MAV_5022|M.avium_104 VA------DLAVGARVVLVFLPRCGRCAACATEGLTPCEPGSAANGAGTL
MUL_4777|M.ulcerans_Agy99 VD------GWSEGDRVVVAAGRPCQSCPNCRRGDTVNCLR----------
MSMEG_2882|M.smegmatis_MC2_155 VHSDSDGRPLEVGTRVVFPYFFCCHTCRNCLAGRRNACVNLKMAMLG---
Mvan_2879|M.vanbaalenii_PYR-1 VTADSNGKPLELGTRVVFPYFFCCHTCRNCLAGRRNACLNLKMAMLG---
Mb1557|M.bovis_AF2122/97 AEQRWG---VSAGDRVAVEVFQSCRQCANCRGGEYRRCVRHGLADMYGFI
Rv1530|M.tuberculosis_H37Rv AEQRWG---VSAGDRVAVEVFQSCRQCANCRGGEYRRCVRHGLADMYGFI
TH_4349|M.thermoresistible__bu VG------TVTEGDAVAVYGPWGCGTCERCMGGAENYCENPAGAPVLS--
. * * . * * * *
Mflv_2766|M.gilvum_PYR-GCK TDG--------TELTPALGIGAFADKTLVHEGQCTKVDPAADP--AAAGL
MLBr_01784|M.leprae_Br4923 IDG--------TELTPALGIGAFADKTLVHSGQCTKVDPAADP--AVAGL
MAB_1869c|M.abscessus_ATCC_199 TDG--------TELTPALGIGAFIEKTLVHAGQCTKVDPTADP--AVVGL
MMAR_0405|M.marinum_M LGGGIRISRSGQPIYHHLGVSGFATHAVVNRASAVAVPDEVPP--QVAAL
MAV_5022|M.avium_104 LGGDIRLSRAGRPVYHHLGVSGFATHAVVNRASAVEVPPDVPA--EVAAL
MUL_4777|M.ulcerans_Agy99 -----------IQLMAFAYDGAWAEYTVAQAAGLTRVPDNVSL--DQAAI
MSMEG_2882|M.smegmatis_MC2_155 -----------RADEPPHFVGGYADYFLLPAKAVVYTVPDSVS--DEIAA
Mvan_2879|M.vanbaalenii_PYR-1 -----------RADEPPYFVGGYADYFVLPAGAVVYTVPDAVS--DDIAA
Mb1557|M.bovis_AF2122/97 -----------PVDREPGLWGGYAEYQYLAPDSMVLRVAGDLS--PEVAT
Rv1530|M.tuberculosis_H37Rv -----------PVDREPGLWGGYAEYQYLAPDSMVLRVAGDLS--PEVAT
TH_4349|M.thermoresistible__bu ------------GGAGLGMDGGMAEYLLVPAERLVVKLPESMTPVTAAAL
.. . .
Mflv_2766|M.gilvum_PYR-GCK LGCGVMAGLGAAINTGGITRDDTVAVIGCGGVGDAAIAGAALVGAKTIIA
MLBr_01784|M.leprae_Br4923 LGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAISGAALVGANRIIA
MAB_1869c|M.abscessus_ATCC_199 LGCGVMAGLGAAVNTGNVGRDDTVAVIGCGGVGDAAIAGARLAGARKIIA
MMAR_0405|M.marinum_M LGCAVLTGGGAVLNVGKPVAGQSVAVVGLGGVGMAAVLTALTQDGVAVFG
MAV_5022|M.avium_104 LGCAVLTGGGAVLNVGRPAPGQTVAVVGLGGVGMAALLTALTYDGVPVVA
MUL_4777|M.ulcerans_Agy99 LADAVSTPFGAVVRTGKVAVGESVGVWGVGGVGTHIVQLARLVGAVPVIA
MSMEG_2882|M.smegmatis_MC2_155 GANCALSQVMYGLERVDLQLGETVVVQGAGALGLYAVAVAKARGAKTVIA
Mvan_2879|M.vanbaalenii_PYR-1 GANCALSQVMYGLERVDQQLGEYVVVQGAGALGLYAVAVAKARGAAKVIA
Mb1557|M.bovis_AF2122/97 LFNPLGAGIRWGVTIPETKPGDVVAVLGPGIRGLCAAAAAKGAGAGFVMV
Rv1530|M.tuberculosis_H37Rv LFNPLGAGIRWGVTIPETKPGDVVAVLGPGIRGLCAAAAAKGAGAGFVMV
TH_4349|M.thermoresistible__bu TDAGLTPYHAIRRSWPKLTPTSTAVVIGVGGLGHVAVQLVKATTAARLIA
. . . * * * * . . :.
Mflv_2766|M.gilvum_PYR-GCK VDTDN---RKLDWARQFGATHTINAKDLD---PVKTIQDLTDGFG--TDV
MLBr_01784|M.leprae_Br4923 VDIDD---TKLEWARTFGATHTVNALELE---VVKTIQDLTSGFG--VDV
MAB_1869c|M.abscessus_ATCC_199 IDTDD---TKLAWAKDFGATDTINARKVDD--VVTAIQELTDEFG--ADV
MMAR_0405|M.marinum_M VDQLP---QKLSAAEALGASATY-----------TPQQAAAAGVK--ADV
MAV_5022|M.avium_104 VDQLP---EKLAAARRLGAHQAY-----------TPQQAVEARVK--AAV
MUL_4777|M.ulcerans_Agy99 LDINP---VVLDRALELGADYAFDSRDDQ---LHDKLAEITGGRM--LDV
MSMEG_2882|M.smegmatis_MC2_155 IDGVP---ERLELAKAFGADEVIDITATTDKDRVKIVRRLTDRQG--ADV
Mvan_2879|M.vanbaalenii_PYR-1 IDGVP---ERLELATAFGADAVIDITATTDKDRAKIVRSLTDGQG--ADV
Mb1557|M.bovis_AF2122/97 TGLGPRDADRLALAAQFGADLAVDVAIDDP------VAALTEQTGGLADV
Rv1530|M.tuberculosis_H37Rv TGLGPRDADRLALAAQFGADLAVDVAIDDP------VAALTEQTGGLADV
TH_4349|M.thermoresistible__bu VDNRS---EALELAREMGADHVVRSDEQ----AVEAIWELTGGRG--ADV
. * * :** . *
Mflv_2766|M.gilvum_PYR-GCK VIDAVG-RPETWKQAFYARDLAGTVVLVGVP-TPDMTLDMPLIDFFSRGG
MLBr_01784|M.leprae_Br4923 VIDTVG-RPETWKQAFYARDLAGTVVLVGVP-TPDMRLDMPLLDFFSHGG
MAB_1869c|M.abscessus_ATCC_199 VIDAVG-RPETWKQAFYARDLAGTVVLVGVP-TPDMTLEMPLIDFFSRGG
MMAR_0405|M.marinum_M VIEAAG-HPAALETAIGLTAPGGRTITVGLP-PPDARISVAPLGFVAEGR
MAV_5022|M.avium_104 VIEAVG-HPAALQTAIDLTAPGGRTITVGLP-PPDARITVSPLGFVAEGR
MUL_4777|M.ulcerans_Agy99 AFDAVG-LKATFEQALGSLTAGG--RLVGVG-TSAESPTVGPTSMFGLTH
MSMEG_2882|M.smegmatis_MC2_155 VVEVVG-HPSAIDEGLKLLGQFGRYVEIGNINIGK-TFEFDPSRFVFANK
Mvan_2879|M.vanbaalenii_PYR-1 VVEVVG-HPSAIDEGLKLLGQFGRYVEIGNINIGR-TFEFDPSRFVFANK
Mb1557|M.bovis_AF2122/97 VVDVTAKAPAAFAQAIALARPAGTVVVAGTRGVGSGAPGFSPDVVVFKEL
Rv1530|M.tuberculosis_H37Rv VVDVTAKAPAAFAQAIALARPAGTVVVAGTRGVGSGAPGFSPDVVVFKEL
TH_4349|M.thermoresistible__bu LLDFVG-ADATIELARAAARPLGDVTIVGIAGGSVPLSFFSQPYEVSIQT
.: .. : . * * . .
Mflv_2766|M.gilvum_PYR-GCK SLKSSWYGDCLPERDFPTLISLYLQGRFPLDKFVSERITLDQIEDAFHKM
MLBr_01784|M.leprae_Br4923 SLKSSWYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVEEAFHKI
MAB_1869c|M.abscessus_ATCC_199 SLKSSWYGDCLPERDFPTLVSLYQQGRLPLDRFVSERIGIESIEAAFEKM
MMAR_0405|M.marinum_M SLIGSYLGSAVPSRDIPRFVSLWQSGRLPVESLVSATIRLDDLNEAMDHL
MAV_5022|M.avium_104 SLIGSYLGSAVPSRDIPRFVELWRAGRLPVESLVSSTIRLEDINEAMDHL
MUL_4777|M.ulcerans_Agy99 KQVLGHLG--YQNVDIETLAKLVSLGRLDLSRSISEIVPLEDLSLGIEKL
MSMEG_2882|M.smegmatis_MC2_155 TMVGVSLYDPAVLSRALTFLAEY-QDRLPFDRLAAACYRLEDINDAFSAA
Mvan_2879|M.vanbaalenii_PYR-1 TMVGVSLYDPSVLSRALAFLEEY-QHRLPFDRLAAARYSLDDINEAFAAA
Mb1557|M.bovis_AF2122/97 RVLGALGVDATAYRAALDLLVS---GRYPFASLPRRCVRLEGAEDLLATM
Rv1530|M.tuberculosis_H37Rv RVLGALGVDATAYRAALDLLVS---GRYPFASLPRRCVRLEGAEDLLATM
TH_4349|M.thermoresistible__bu TYWG----------TRPELVELLDLAARGLVHVESTTYPLDDALQAYRDL
: . :
Mflv_2766|M.gilvum_PYR-GCK HAGSDD-VVLRSVVVL--
MLBr_01784|M.leprae_Br4923 HGG----KVLRSVVML--
MAB_1869c|M.abscessus_ATCC_199 HHG----EVLRSVVVL--
MMAR_0405|M.marinum_M ADG----TAVRQIIVF--
MAV_5022|M.avium_104 ADG----TAVRQLIRFDA
MUL_4777|M.ulcerans_Agy99 ERQ-DG-NPIRILVQP--
MSMEG_2882|M.smegmatis_MC2_155 ENKRD----VRASIVP--
Mvan_2879|M.vanbaalenii_PYR-1 EDKRD----VRASIVP--
Mb1557|M.bovis_AF2122/97 AGERDGVPPIHGVLTP--
Rv1530|M.tuberculosis_H37Rv AGERDGVPPIHGVLTP--
TH_4349|M.thermoresistible__bu RAGRVR---GRAVIVP--
: :