For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NDSPIVVNHHADHPGFSGVTGAIFGVLFVALGRANAKVAIEATQVGPIDHVVDVGCGPGTAARMAARRGA RVTAVDPSAEMLRVGRLVTPSAAGKNITWRPGTAEDVPVDDGTATVVWALATVHHWQDVVRALQQANRVL APGGRLLAIERQVARGATGFASHGWTREQADAFATLCRDAGFGEVEVSERPRGRKSVWLVSAIKR*
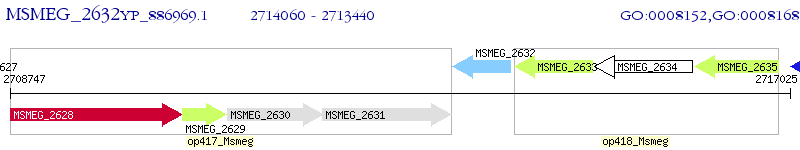
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2632 | - | - | 100% (206) | SAM-dependent methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1650 | - | 1e-10 | 38.54% (96) | methyltransferase type 11 |
| M. smegmatis MC2 155 | MSMEG_2285 | - | 2e-10 | 35.34% (133) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3374 | - | 5e-13 | 41.67% (96) | methyltransferase (methylase) |
| M. gilvum PYR-GCK | Mflv_4035 | - | 1e-58 | 54.04% (198) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3342 | - | 5e-13 | 41.67% (96) | methyltransferase (methylase) |
| M. leprae Br4923 | MLBr_01723 | - | 6e-09 | 33.01% (103) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_3689 | - | 1e-11 | 40.62% (96) | putative methyltransferase |
| M. marinum M | MMAR_1183 | - | 4e-12 | 40.62% (96) | methyltransferase |
| M. avium 104 | MAV_4317 | - | 4e-12 | 40.62% (96) | putative methyltransferase |
| M. thermoresistible (build 8) | TH_1968 | - | 4e-12 | 40.62% (96) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_1436 | - | 3e-12 | 40.62% (96) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_2319 | - | 7e-56 | 59.22% (179) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4035|M.gilvum_PYR-GCK ----------------MDGSERPVNHHGDHPGFSGLTGQLCAVAFLLTGR
Mvan_2319|M.vanbaalenii_PYR-1 ------------------------------------------MLFLLAGR
MSMEG_2632|M.smegmatis_MC2_155 ----------------MNDSPIVVNHHADHPGFSGVTGAIFGVLFVALGR
Mb3374|M.bovis_AF2122/97 ---------------------MTCSRRDMSLSFG-----SAVGAYERGRP
Rv3342|M.tuberculosis_H37Rv ---------------------MTCSRRDMSLSFG-----SAVGAYERGRP
MMAR_1183|M.marinum_M ---------------------MSGSAQDPSLSFG-----SAAAAYERGRP
MUL_1436|M.ulcerans_Agy99 --------------------MVSGSAQDPSLSFG-----SAAAAYERGRP
MAV_4317|M.avium_104 ---------------------MTRSRHDRSLSFG-----SAAAAYERGRP
TH_1968|M.thermoresistible__bu ------------------------------LSFG-----AEAAAYERGRP
MAB_3689|M.abscessus_ATCC_1997 ---------------------MPDAQAEPSLSFG-----SQAAAYERGRP
MLBr_01723|M.leprae_Br4923 MTRSTEISANAVPNPHATAEQVAAARKDSKLAQVLYHDWEAENYDEKWSI
Mflv_4035|M.gilvum_PYR-GCK ETGRLAADLT-------------AVSATDHVLDIGCGPGNACRIAAARGA
Mvan_2319|M.vanbaalenii_PYR-1 RTAGLAAALT-------------EVTASDHVVDIGCGPGNGARIAAQRGA
MSMEG_2632|M.smegmatis_MC2_155 ANAKVAIEAT-------------QVGPIDHVVDVGCGPGTAARMAARRGA
Mb3374|M.bovis_AF2122/97 SYPPEAIDWL-------------LPAAARRVLDLGAGTGKLTTRLVERGL
Rv3342|M.tuberculosis_H37Rv SYPPEAIDWL-------------LPAAARRVLDLGAGTGKLTTRLVERGL
MMAR_1183|M.marinum_M SYPPEAIDWL-------------LPAGARDVLDLGAGTGKLTTRLVERGL
MUL_1436|M.ulcerans_Agy99 SYPPEAIDWL-------------LPAGARDVLDLGAGTGKLTTRLVERGL
MAV_4317|M.avium_104 SYPPEAIDWL-------------LPVGARQVLDLGAGTGKLTTRLVERGL
TH_1968|M.thermoresistible__bu SYPPEAIDWL-------------LPSDARDVLDLGAGTGKLTTRLVERGL
MAB_3689|M.abscessus_ATCC_1997 SYPPEAIDWL-------------LPPGAHDVLDLGAGTGKLTTRLVERGL
MLBr_01723|M.leprae_Br4923 SYDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLIQSGV
. .:::*.*.* *
Mflv_4035|M.gilvum_PYR-GCK RVTG--VDPSAAMLRVARIVTHG---ASVTWVRGGAEALPVPDAGATVAW
Mvan_2319|M.vanbaalenii_PYR-1 RVTG--VDPSRSMLRVARAVTRGR--PAITWAEGTAEALPVPDASATVVW
MSMEG_2632|M.smegmatis_MC2_155 RVTA--VDPSAEMLRVGRLVTPSAAGKNITWRPGTAEDVPVDDGTATVVW
Mb3374|M.bovis_AF2122/97 DVVA--VDPIPEMLDVLRAALPQT-----VALLGTAEEIPLDDN-SVDAV
Rv3342|M.tuberculosis_H37Rv DVVA--VDPIPEMLDVLRAALPQT-----VALLGTAEEIPLDDN-SVDAV
MMAR_1183|M.marinum_M DVVA--VDPIPEMLEVLRASLPRT-----LALLGTAEEIPLEDN-SVDAV
MUL_1436|M.ulcerans_Agy99 DVVA--VDPIPEMLEVLRASLPRT-----LALLGTAEEIPLEDN-SVDAV
MAV_4317|M.avium_104 DVVA--VDPIPDMLEVLRSSLPET-----RALLGTAEEIPLEDN-SVDVV
TH_1968|M.thermoresistible__bu NVTA--VDPIPEMLELLSKSLPDT-----PALLGTAEEIPLPDN-SVDAV
MAB_3689|M.abscessus_ATCC_1997 NVVA--VDPLAEMLEVLSNSLPDT-----PALLGTAEEIPLPDN-SVDAV
MLBr_01723|M.leprae_Br4923 ARRGSVTDLSPGMVKIATRNGQSLG-LDINGRVADAEGIPYDDNTFDLVV
. .* *: : . ** :* * .
Mflv_4035|M.gilvum_PYR-GCK ALATVHHWPDVDAGLAEIHRALGPGARF--------------LAVERQTE
Mvan_2319|M.vanbaalenii_PYR-1 ALATVHHWRDVGAGLSEIHRVLVPGGRL--------------LAVERQVA
MSMEG_2632|M.smegmatis_MC2_155 ALATVHHWQDVVRALQQANRVLAPGGRL--------------LAIERQVA
Mb3374|M.bovis_AF2122/97 LVAQAWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGEIIG
Rv3342|M.tuberculosis_H37Rv LVAQAWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGEIIG
MMAR_1183|M.marinum_M LVAQAWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGRIIG
MUL_1436|M.ulcerans_Agy99 LVAQAWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGRIIG
MAV_4317|M.avium_104 LVAQAWHWVDPERAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGRIIG
TH_1968|M.thermoresistible__bu LVAQAWHWFDVDRAVREVARVLRPGGRLGLVWNTRDERMGWVKELGRIIG
MAB_3689|M.abscessus_ATCC_1997 LVAQAWHWVDPERAIPEVARVLRPGGRLGLVWNVRDERLGWVSDLGHIIG
MLBr_01723|M.leprae_Br4923 GHAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTDVGNRYARVFADLTW
* * * .: : *.* **.*: .
Mflv_4035|M.gilvum_PYR-GCK ------PDATGISSHG----------------WTG----AQAETFAAMCE
Mvan_2319|M.vanbaalenii_PYR-1 ------PDATGFASHG----------------WTG----QQAETFAALCV
MSMEG_2632|M.smegmatis_MC2_155 ------RGATGFASHG----------------WTR----EQADAFATLCR
Mb3374|M.bovis_AF2122/97 ------RDGDPVRDRV-TLPEPFTTVQRHQVEWTNYL--TPQALIDLVAS
Rv3342|M.tuberculosis_H37Rv ------RDGDPVRDRV-TLPEPFTTVQRHQVEWTNYL--TPQALIDLVAS
MMAR_1183|M.marinum_M ------RDSDPVRDKV-TLPEPFTGVARHHVEWTNYL--TPQALIDLVAS
MUL_1436|M.ulcerans_Agy99 ------RDSDPVRDKV-TLPEPFTGVARHHVEWTNYL--TPQALIDLVAS
MAV_4317|M.avium_104 ------SDGD-GRTHV-ALPEPFTDLAHHEVEWTNYL--TPQALIDLVAS
TH_1968|M.thermoresistible__bu ------DEHDPLSKTV-TLPEPFTDVQRHQVEWTNYL--TPQALIDLVAS
MAB_3689|M.abscessus_ATCC_1997 ------HENAADSADVGALPAPFTGQERHQVEWTNYL--TPQALLDMVAS
MLBr_01723|M.leprae_Br4923 KVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVLETMAT
*. : :.
Mflv_4035|M.gilvum_PYR-GCK SAGLTTVRVGSGRAGKRRVWTVHATRP-----------------------
Mvan_2319|M.vanbaalenii_PYR-1 TAGLADAKVNADRAGRREVWTVHAVRP-----------------------
MSMEG_2632|M.smegmatis_MC2_155 DAGFGEVEVSERPRGRKSVWLVSAIKR-----------------------
Mb3374|M.bovis_AF2122/97 RSYCITSPAQVRTKTLDRVRQLLATHPALANSNGLALPYVTVCVRATLA-
Rv3342|M.tuberculosis_H37Rv RSYCITSPAQVRTKTLDRVRQLLATHPALANSNGLALPYVTVCVRATLA-
MMAR_1183|M.marinum_M RSYCITSPAEVRTKTLDQVRELLATHPALANSAGLALPYVTVCVSATLAA
MUL_1436|M.ulcerans_Agy99 RSYCITSPAEVRTKTLDQVRELLATHPALANSAGLALPYVTVCVSATLAA
MAV_4317|M.avium_104 RSYCITSPTEVRTRTLDQVRHLLATHPALANSTGLALPYVTRCTRATLAG
TH_1968|M.thermoresistible__bu RSYCITSPAKVRARTLERVRELLATHPALANNSGLALPYITVCIRATLA-
MAB_3689|M.abscessus_ATCC_1997 RSYCITAPEAARKKTLQQVRELLATHPALAQSAGLALPYVTVSIRATLNR
MLBr_01723|M.leprae_Br4923 NAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFTSWKTL
: :
Mflv_4035|M.gilvum_PYR-GCK ----------------------------
Mvan_2319|M.vanbaalenii_PYR-1 ----------------------------
MSMEG_2632|M.smegmatis_MC2_155 ----------------------------
Mb3374|M.bovis_AF2122/97 ----------------------------
Rv3342|M.tuberculosis_H37Rv ----------------------------
MMAR_1183|M.marinum_M ----------------------------
MUL_1436|M.ulcerans_Agy99 ----------------------------
MAV_4317|M.avium_104 ----------------------------
TH_1968|M.thermoresistible__bu ----------------------------
MAB_3689|M.abscessus_ATCC_1997 ----------------------------
MLBr_01723|M.leprae_Br4923 SWVDANVWRRVIPKSWFYNIIITGVKPS