For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSMRDAVICEPVRTPIGRYGGMFRSLTAVDLGVTALKGLLERTGIAADQVEDVILGHCYPNSEAPAIGRV VALDAGLPITVPGMQVDRRCGSGLQAVIQACLQVRSGDHDLVVAGGAESMSNVAFYSTDMRWGGARTGVQ IHDGLARGRTTAGGKFHPVPGGMLETAENLRREYHISRTEQDELAVRSHQRAVAAQSEGVLAEEIIPVPV RTRDGEETISVDEHPRADTTVEALAKLKPVLLKQDPEATVTAGNSSGQNDAASMCIVTTPEKAAELGLKP LVRLVSWGSAGVAPDLMGIGPVPATEVALAKAGLTLADIDLIELNEAFAAQALAVMREWKFGEADHERTN VRGSGISLGHPVGATGGRMLATLARELHRREARYGLETMCIGGGQGLAAVFERVQEG
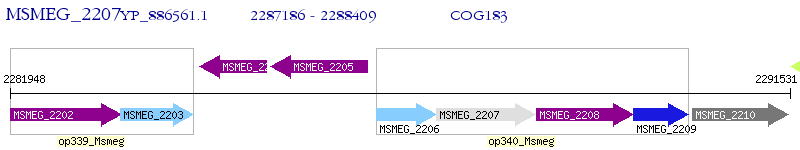
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2207 | - | - | 100% (407) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_1896 | - | e-137 | 61.33% (406) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_3012 | - | 1e-68 | 40.39% (411) | acetyl-CoA acetyltransferases |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1103c | fadA3 | 3e-67 | 38.61% (417) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_4368 | - | 0.0 | 81.59% (402) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv1074c | fadA3 | 3e-67 | 38.61% (417) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_02162 | fadA | 1e-59 | 38.03% (426) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_0902 | - | 3e-67 | 39.85% (409) | beta-ketoadipyl-CoA thiolase |
| M. marinum M | MMAR_4032 | - | 0.0 | 82.88% (403) | beta-ketoacyl CoA thiolase |
| M. avium 104 | MAV_1573 | - | 0.0 | 84.86% (403) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_0255 | - | 4e-64 | 39.26% (405) | acetyl-CoA acetyltransferase |
| M. ulcerans Agy99 | MUL_0207 | fadA3 | 1e-67 | 39.57% (422) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1976 | - | 0.0 | 80.85% (402) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1103c|M.bovis_AF2122/97 ----MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQMVRAALDKVPALN
Rv1074c|M.tuberculosis_H37Rv ----MPEAVIVSTARSPIGRAMKGSLVGMRPDDLAVQMVRAALDKVPALN
MUL_0207|M.ulcerans_Agy99 ----MPEAVIVSTARSPIGRAMKASLVSMRPDDLAVQMVRAALDKVPALN
MMAR_4032|M.marinum_M MSAAMRQAVICEPVRTPIGR-YGGMFKSHSAVDLGVVALRGLLDRT-GLA
MAV_1573|M.avium_104 -MSAVREAVICEPVRTPIGR-YGGMFKALTAVDLGVAALGGLLQRT-GLP
MSMEG_2207|M.smegmatis_MC2_155 --MSMRDAVICEPVRTPIGR-YGGMFRSLTAVDLGVTALKGLLERT-GIA
Mflv_4368|M.gilvum_PYR-GCK ---MTREAVICEPVRTPIGR-YNGMFKSLTAVELGVVALKGLLERT-QVA
Mvan_1976|M.vanbaalenii_PYR-1 ---MMRDAVVCEPVRTPIGR-YNGMFKSLTAVELGVAALQGLLART-GVA
MLBr_02162|M.leprae_Br4923 ---MSEEAFIYEAIRTPRGKQKNGSLHEVKPVSLVVGLVEELRKRNPDLD
TH_0255|M.thermoresistible__bu ----MRETVIVEAVRTAVGK-RNGALADVHAADLSAVVLSELVER-AGID
MAB_0902|M.abscessus_ATCC_1997 ---MVEQVFVVDGVRTAQGR-YGGALASVRPDDLAARVIAEVVAR-AGVP
:..: . *:. *: . : . .* . : : :
Mb1103c|M.bovis_AF2122/97 PHQIDDLMMGCGLPGGESGFNIARVVAVALGYDF-LPGTTVNRYCSSSLQ
Rv1074c|M.tuberculosis_H37Rv PHQIDDLMMGCGLPGGESGFNIARVVAVALGYDF-LPGTTVNRYCSSSLQ
MUL_0207|M.ulcerans_Agy99 PHQIDDLIMGCGLPGGESGFNIGRVVAVALGYDF-LPGTTVNRYCSSSLQ
MMAR_4032|M.marinum_M PAAVGDVILGHCYPSSE-APAIGRVVALDAGLPVTVPGMQVDRRCGSGLQ
MAV_1573|M.avium_104 PEAVQDVILGHCYPSSE-APAIGRVVALDSGLPVTVPGMQVDRRCGSGLQ
MSMEG_2207|M.smegmatis_MC2_155 ADQVEDVILGHCYPNSE-APAIGRVVALDAGLPITVPGMQVDRRCGSGLQ
Mflv_4368|M.gilvum_PYR-GCK PEAIEDVIVGHCYPSME-APAIGRVIALDAGLPVTVPGMQLDRRCGSGLQ
Mvan_1976|M.vanbaalenii_PYR-1 PGAVEDVVVGHCYPSME-APAIGRVIALDAGLPVTVPGMQIDRRCGSGLQ
MLBr_02162|M.leprae_Br4923 ENLISDVILGCVSPVGDQGGDIARATVLAAGLPVTAGGVQLNRFCASGLE
TH_0255|M.thermoresistible__bu PELIDDVIWGCVSQVGDQSSNIGRYAVLAAGWPETIPGTTVNRACGSSQQ
MAB_0902|M.abscessus_ATCC_1997 ADAIGEVILGAANQAGEDNRNVARMAALLAGLPNTVPGYTVNRLCASGLT
: ::: * : :.* .: * * ::* *.*.
Mb1103c|M.bovis_AF2122/97 TTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDS-WPDTKNP-------
Rv1074c|M.tuberculosis_H37Rv TTRMAFHAIKAGEGDAFISAGVETVSRFAKGNSDS-WPDTKNP-------
MUL_0207|M.ulcerans_Agy99 ITRVAFHAIKAGEGDAFISAGVETVSRFGKGNSDS-LPDTKNP-------
MMAR_4032|M.marinum_M AVIQACQQVSSGDSDLVVAGGCESMSNVTFYSTDMRWGGARGG-------
MAV_1573|M.avium_104 AVIQACLQVRNGDHDLVIAGGCESMSNVAFYSTDMRWGGARSG-------
MSMEG_2207|M.smegmatis_MC2_155 AVIQACLQVRSGDHDLVVAGGAESMSNVAFYSTDMRWGGARTG-------
Mflv_4368|M.gilvum_PYR-GCK AVIQAAMQVASGTNDLVVAGGAESMSNVVFHSTDMRWGGARTG-------
Mvan_1976|M.vanbaalenii_PYR-1 AVIQAAMQVASGNNDLVVAGGAESMSNVVFHSTDMRWGGARGG-------
MLBr_02162|M.leprae_Br4923 AVNTAAQKVRSGWDDLVLAGGVESMSRVPMGSD-----------------
TH_0255|M.thermoresistible__bu ALDFAVHAVMSGQQDVVVAGGVEVMSRVPLGAAR----------------
MAB_0902|M.abscessus_ATCC_1997 AISSAAQAIRAGEADIVVAGGVESMTRAPWVMAKPSAPWARPGEVVDTSL
* : * * .::.* * ::.
Mb1103c|M.bovis_AF2122/97 -----LFDGAQERSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIM
Rv1074c|M.tuberculosis_H37Rv -----LFDGAQERSAAAAAGADEWHDPRTDQKLPDIYIAMGQTAENVAIM
MUL_0207|M.ulcerans_Agy99 -----LFDEAQERTAAAAAGAEEWHDPRADGNLPDVYIAMGQTAENVAII
MMAR_4032|M.marinum_M -----IRVHDGLARGRVTAGGRHYPVPG----------GMLETAENLRRQ
MAV_1573|M.avium_104 -----VRVHDGLARGRTTAGGRRYPVPG----------GMLETAENLRRQ
MSMEG_2207|M.smegmatis_MC2_155 -----VQIHDGLARGRTTAGGKFHPVPG----------GMLETAENLRRE
Mflv_4368|M.gilvum_PYR-GCK -----ITVHDALARGRTTAGGKHYPVPG----------GMLETAENLRRE
Mvan_1976|M.vanbaalenii_PYR-1 -----ITVHDALARGRTTAGGKNYPVPG----------GMLETAENLRRD
MLBr_02162|M.leprae_Br4923 ------------GGAMGLDPATNYDVMF---------VPQGIGADLIATI
TH_0255|M.thermoresistible__bu ----------ATGMPYGPKVLERYDNFG---------FNQGISAELIAER
MAB_0902|M.abscessus_ATCC_1997 GWRFTNPRFAQMDAGIAADAGPETVKVT---------LAMGETAEEVAVA
*: :
Mb1103c|M.bovis_AF2122/97 TGISREEQDRWGVRSQNRAEEAIKNGFFEREITPVTLPDG---TTVSTDD
Rv1074c|M.tuberculosis_H37Rv TGISREEQDRWGVRSQNRAEEAIKNGFFEREITPVTLPDG---TTVSTDD
MUL_0207|M.ulcerans_Agy99 SGISREDQDHWGVRSQNRAEEAIKGGFFEREISPVTLPDG---TTVSTND
MMAR_4032|M.marinum_M YGISRLEQDELAVTSHQRAVAAQKDGIFAEETIAVPVRTRQGEELIDTDE
MAV_1573|M.avium_104 YGISRQEQDELAVRSHQRAVAAQKDGILAQEIIAVAVPTRHGDELIDTDE
MSMEG_2207|M.smegmatis_MC2_155 YHISRTEQDELAVRSHQRAVAAQSEGVLAEEIIPVPVRTRDGEETISVDE
Mflv_4368|M.gilvum_PYR-GCK YGISREEQDELAVRSHSKAVRAQQDGLLAETIIPVTVSGRSGDAVIDVDE
Mvan_1976|M.vanbaalenii_PYR-1 YGISRQEQDELAVLSHRRAVTAQKDGLLAETIIPVTVTSRSGDQVIDTDE
MLBr_02162|M.leprae_Br4923 EGFSRDDVDVYALRSQEKAAAAWSGGYFAKSVVPVRDQNGL--LILDHDE
TH_0255|M.thermoresistible__bu WNLSRTRLDEYSAQSHERAAAAQSSGAFKDQIVPVFTDD----AVVTEDE
MAB_0902|M.abscessus_ATCC_1997 EGISREDSDAFALRSHVRALAAQDAGRFVKEIVPIETVDG----VVDTDE
:** * . *: :* * . * : .: : ::
Mb1103c|M.bovis_AF2122/97 GPRPGTTYEKVSELKPAFR---------------------PNGTVTAGNA
Rv1074c|M.tuberculosis_H37Rv GPRPGTTYEKVSELKPAFR---------------------PNGTVTAGNA
MUL_0207|M.ulcerans_Agy99 GPRAGTTYEKVSQLKPVFR---------------------PNGTVTAGNA
MMAR_4032|M.marinum_M HPRADTSVEALSGLKPILHQ------------------DDPEATVTAGNA
MAV_1573|M.avium_104 HPRADTSVESLSKLKPVLLE------------------QDPEATVTAGNA
MSMEG_2207|M.smegmatis_MC2_155 HPRADTTVEALAKLKPVLLK------------------QDPEATVTAGNS
Mflv_4368|M.gilvum_PYR-GCK HPRADTSPESLAKLKPVLAA------------------SDPEATVTAGNS
Mvan_1976|M.vanbaalenii_PYR-1 HPRADTSLESLAKLKPVLGK------------------SDPDATVTAGNS
MLBr_02162|M.leprae_Br4923 HMRPDTSKEGLAKLKPAFEGLAAMGGFDDVALQKYHWVEKINHVHTGGNS
TH_0255|M.thermoresistible__bu GIRRGTTVEKLATLKPAFT---------------------ENGVIHAGNS
MAB_0902|M.abscessus_ATCC_1997 GPRRQTTLAKLAALRPVFR---------------------AGGIVTAGSS
* *: :: *:* : .*.:
Mb1103c|M.bovis_AF2122/97 CPLNDGAAAVVITSDTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAS
Rv1074c|M.tuberculosis_H37Rv CPLNDGAAAVVITSDTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAS
MUL_0207|M.ulcerans_Agy99 CPLNDGAAAVVITSDTKAKELGLTPLARIVSTGVSGLSPEIMGLGPIEAC
MMAR_4032|M.marinum_M SGQNDAASVCVVTTPEKADEYGLRPLVRLVSWGLAGVAPDVMGIGPVPAT
MAV_1573|M.avium_104 SGQNDAASMCVVTTPEKADEHGLRPLVRLVSWAVAGVAPHIMGIGPVPAT
MSMEG_2207|M.smegmatis_MC2_155 SGQNDAASMCIVTTPEKAAELGLKPLVRLVSWGSAGVAPDLMGIGPVPAT
Mflv_4368|M.gilvum_PYR-GCK SGQNDAASMCLVTTPEKAAELGLVPLVRIVSWGVAGVAPKIMGIGPVPAT
Mvan_1976|M.vanbaalenii_PYR-1 SGQNDAASMCLVTTPQKAAELGLRPLVRLVSWGVAGVAPNVMGIGPVPAT
MLBr_02162|M.leprae_Br4923 SGIVDGAALVMIGSEAVGKSQGLTPRARMVATATSGADPVIMLTGPTPAT
TH_0255|M.thermoresistible__bu SQISDGAAALLVMTAETAVTLGLTPIVRYRAGAVTGADPVLMLTGPIPAT
MAB_0902|M.abscessus_ATCC_1997 SPLSDGAAAVVLAGERAVRQHGLTVRARVVAAASAGVAPNLMGLGPVPAV
. *.*: :: ** .* : . :* * :* ** *
Mb1103c|M.bovis_AF2122/97 KKALERAGMAITDIDLVEINEAFAVQVLGSARELG---IDEDKLNISGGA
Rv1074c|M.tuberculosis_H37Rv KKALERAGMAITDIDLVEINEAFAVQVLGSARELG---IDEDKLNISGGA
MUL_0207|M.ulcerans_Agy99 KKALQRAGMSISDIDLVEINEAFAVQVLGSARELG---IDEDELNVSGGA
MMAR_4032|M.marinum_M EAALAKAALRLADIDLIELNEAFAAQALAVMREWNFGAADRDRTNVHGSG
MAV_1573|M.avium_104 EVALAKAGLSLSDIDLIELNEAFAAQALAVMREWKFGAADHDRTNVHGSG
MSMEG_2207|M.smegmatis_MC2_155 EVALAKAGLTLADIDLIELNEAFAAQALAVMREWKFGEADHERTNVRGSG
Mflv_4368|M.gilvum_PYR-GCK EVALRRAGLQLPDIDLIELNEAFAAQALACLREWKFTETDLERTNVHGSG
Mvan_1976|M.vanbaalenii_PYR-1 EVALGNAGLALSDVDLIELNEAFAAQALACMREWKFTQADLERTNVHGSG
MLBr_02162|M.leprae_Br4923 RKVLDRAGLTVDDIDLFELNEAFASVVLKFQKDLN---IPDEKLNVNGGA
TH_0255|M.thermoresistible__bu EKVLRKTGLNIDDIGVFEVNEAFAPVPLAWQAETG---ADPAKLNPLGGA
MAB_0902|M.abscessus_ATCC_1997 RKVVDRVGWRVDAIDATEINEAFAAQTLAVLRRLG---LDEDTVNADGGA
. .: ... : :. *:***** * * *..
Mb1103c|M.bovis_AF2122/97 IALGHPFGMTGARITTTLLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA
Rv1074c|M.tuberculosis_H37Rv IALGHPFGMTGARITTTLLNNLQTYDKTFGLETMCVGGGQGMAMVIERLA
MUL_0207|M.ulcerans_Agy99 IALGHPFGMTGARITTTLLNNLQTHDKTFGLETMCVGGGQGMAMVIERLS
MMAR_4032|M.marinum_M ISLGHPVGATGARMLATLARELTRRQARYGLETMCIGGGQGLAAIFERVG
MAV_1573|M.avium_104 ISLGHPVGATGGRMLATLARELTRRQARYGLETMCIGGGQGLAAVFERVA
MSMEG_2207|M.smegmatis_MC2_155 ISLGHPVGATGGRMLATLARELHRREARYGLETMCIGGGQGLAAVFERVQ
Mflv_4368|M.gilvum_PYR-GCK ISLGHPVGATGGRMLATLARELVRRDGRFGLETMCIGGGQGLAAVFERVE
Mvan_1976|M.vanbaalenii_PYR-1 ISLGHPVGATGGRMLATLARELNRRDGRFGLETMCIGGGQGLAAVFERVT
MLBr_02162|M.leprae_Br4923 IAMGHPLGATGAMILGTMVDELERCNARRALVTLCIGAGMGVATIIERV-
TH_0255|M.thermoresistible__bu IALGHPLGASGAVLMTRMVHHMRANGIRYGLQTMCEGGGTANATVVELLA
MAB_0902|M.abscessus_ATCC_1997 IALGHPLGCSGARLIVTLLNRLEREGASRGLATLCVGVGQGVAILIEAP-
*::***.* :*. : : .: .* *:* * * . * :.*
Mb1103c|M.bovis_AF2122/97 --
Rv1074c|M.tuberculosis_H37Rv --
MUL_0207|M.ulcerans_Agy99 --
MMAR_4032|M.marinum_M --
MAV_1573|M.avium_104 --
MSMEG_2207|M.smegmatis_MC2_155 EG
Mflv_4368|M.gilvum_PYR-GCK G-
Mvan_1976|M.vanbaalenii_PYR-1 P-
MLBr_02162|M.leprae_Br4923 --
TH_0255|M.thermoresistible__bu --
MAB_0902|M.abscessus_ATCC_1997 --