For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEELTVGEVAKRFRITVRTLHHYDEIGLLSPSRRVTSGYRVYTPADLTRLSQIIVYRRLEFSLDEIASLL DEGDAVSHLLRQRERVMARLGEMKDLVAAIDQALEKAMTNTPMTDDDMRELFGEGFDDYQAEAERKFGDT AEWKESQRRTKSYSKGQWIQLKDEGEAVEKALADAFRGGLAPDSEVAMDAAEQHRLHVNRWFYDCPPDFH RKLGDMYVSDPAYVARYDESFGLPGLAAFCRDAIHANADRAGGDVTG
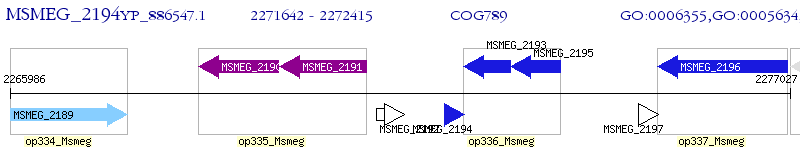
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2194 | - | - | 100% (257) | MerR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1447 | - | 6e-13 | 48.53% (68) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_3180 | - | 7e-09 | 36.89% (103) | transcriptional regulator, MerR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3367 | - | 4e-07 | 31.40% (121) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0148 | - | 1e-117 | 80.08% (251) | MerR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3334 | - | 4e-07 | 31.40% (121) | MerR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4723c | - | 2e-06 | 24.07% (216) | MerR family transcriptional regulator |
| M. marinum M | MMAR_0009 | - | 1e-119 | 80.24% (253) | transcriptional regulator |
| M. avium 104 | MAV_3222 | - | 1e-07 | 40.70% (86) | transcriptional regulator, MerR family protein |
| M. thermoresistible (build 8) | TH_2460 | - | 6e-06 | 28.57% (91) | PUTATIVE putative transcriptional regulator, MerR family |
| M. ulcerans Agy99 | MUL_0011 | - | 1e-120 | 80.63% (253) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1919 | - | 1e-128 | 86.11% (252) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3367|M.bovis_AF2122/97 ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
Rv3334|M.tuberculosis_H37Rv ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
MMAR_0009|M.marinum_M ---------------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRA
MUL_0011|M.ulcerans_Agy99 ---------------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRA
Mvan_1919|M.vanbaalenii_PYR-1 MTSGTTLTYVTQEMQVDELTVGEVAERFGITVRTLHHYDEIGLLTPGRRC
Mflv_0148|M.gilvum_PYR-GCK ---------MTQETRMEELTVGEVAKRFGITIRTLHHYDHIGLLTPEQRN
MSMEG_2194|M.smegmatis_MC2_155 ---------------MEELTVGEVAKRFRITVRTLHHYDEIGLLSPSRRV
TH_2460|M.thermoresistible__bu -----------------------------------------------VRT
MAV_3222|M.avium_104 -----------------MLTIGEVARRSGVAATTLRYYEQIGLLPAPTR-
MAB_4723c|M.abscessus_ATCC_199 -------MATDAEGVAVGWSTRELADLAGTSLRTVRHYHDVGLLGEPERR
*
Mb3367|M.bovis_AF2122/97 ASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILA----IHD-------
Rv3334|M.tuberculosis_H37Rv ASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILA----IHD-------
MMAR_0009|M.marinum_M ASGYRVYTSADLTRLSQVIVYRRLELPLDEIARLLD----EGNAVSHLVR
MUL_0011|M.ulcerans_Agy99 ASGYRVYTSADLTRLSQVIVYRRLELPLDEIARLLD----EGNAVSHLVR
Mvan_1919|M.vanbaalenii_PYR-1 ASGYRVYTPADLTRLSQVIVYRRLDFSLDEIASLLD----EGNEVSHLIR
Mflv_0148|M.gilvum_PYR-GCK ASGYRVYTSADLTRLSQIIVYRRLEMSLDEIASVLD----EGDEISHLRR
MSMEG_2194|M.smegmatis_MC2_155 TSGYRVYTPADLTRLSQIIVYRRLEFSLDEIASLLD----EGDAVSHLLR
TH_2460|M.thermoresistible__bu AAGYRGDTDAAIERLHLIPVYRSAGLPLEEIRALLDG---AADPLARLQR
MAV_3222|M.avium_104 LGGQRRYDEAVLSRLEVIALCKTAGFALDEIQRLFA------DDAPGRPA
MAB_4723c|M.abscessus_ATCC_199 PNGYKSYGVAHLVRVLQIKRLTGLGLSLKQIAEMDEADENQQEALRALDA
* : : *: : :.*.:: : :
Mb3367|M.bovis_AF2122/97 ---RGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDH--ASYGP
Rv3334|M.tuberculosis_H37Rv ---RGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDH--ASYGP
MMAR_0009|M.marinum_M QRERVMARLDEMKDLVEAIDQALEKAMTNTPMTDDDMRELFGDGFDDYRA
MUL_0011|M.ulcerans_Agy99 QRERVMARLDEMKDLVEAIDLALEKAMTNTPMTDDDMRELFGDGFDDYRV
Mvan_1919|M.vanbaalenii_PYR-1 QRERVMSRLDEMKDLVAAIDQALEKAMTNTPMTDDDMRELFGDGFDDYQA
Mflv_0148|M.gilvum_PYR-GCK QRERVMSRLDELTELVGAIDQALENAMTNTPMTDDDMRELFGDSFDDYQA
MSMEG_2194|M.smegmatis_MC2_155 QRERVMARLGEMKDLVAAIDQALEKAMTNTPMTDDDMRELFGEGFDDYQA
TH_2460|M.thermoresistible__bu QHALLLEQADRLQHTIKAVEELMNAHRHGIQLSAEEQVEIFGS--AVFR-
MAV_3222|M.avium_104 SRALARAKLAQIDAQLDSLTRARAVIEWGMRCTCPSIDACTCGIHPTRPA
MAB_4723c|M.abscessus_ATCC_199 ELAATIDRLQGVRDELAQIFSSEIAADLPREFGAAAVDMNFSETDRSLAF
: :
Mb3367|M.bovis_AF2122/97 PTEHDHSTVCWILES----------------------------------D
Rv3334|M.tuberculosis_H37Rv PTEHDHSTVCWILES----------------------------------D
MMAR_0009|M.marinum_M ETEQKWGETTEWKESQRRTRSYGKDEWVRIKAEGQAVEKTLSDAFRAGLP
MUL_0011|M.ulcerans_Agy99 ETEQKWGETAEWKESQRRTKSYGKDEWVRIKAEGQAIEKALSDAFRAGLP
Mvan_1919|M.vanbaalenii_PYR-1 EAEQKWGETAEWKESQRRTKSYGKDEWVQIKSEGEAVEKALSDAFRAGLP
Mflv_0148|M.gilvum_PYR-GCK EAEQKWGETAEWKESQRRATSYGKDDWVRIKAEGQAVEKALADAFRAGLP
MSMEG_2194|M.smegmatis_MC2_155 EAERKFGDTAEWKESQRRTKSYSKGQWIQLKDEGEAVEKALADAFRGGLA
TH_2460|M.thermoresistible__bu --------------------------------------------------
MAV_3222|M.avium_104 PVGPDVRSARRF--------------------------------------
MAB_4723c|M.abscessus_ATCC_199 VMSRVLGPAGLQAFVG-----------MFLDQQDRALDHELENLPADAGE
Mb3367|M.bovis_AF2122/97 LDEPTAIEVSDIHA------------------------------------
Rv3334|M.tuberculosis_H37Rv LDEPTAIEVSDIHA------------------------------------
MMAR_0009|M.marinum_M ADSEEAMNAAEQHRRHVNRWFYDCPPAFHRSLGDMYVSDPRYIATYDESF
MUL_0011|M.ulcerans_Agy99 ADSEEAMNAAEQHRRHVNRWFYDCPPAFHRNLGDMYVSDPRYIATYDESF
Mvan_1919|M.vanbaalenii_PYR-1 PDSAEAMNAAEQHRLHVNRWFYDCPPAFHRKLGDMYVSDPRYVATYDESF
Mflv_0148|M.gilvum_PYR-GCK ADSEEAMNAAEQHRHHVNRWFYDCPPAFHRTLGDMYVSDPRYVATYDESF
MSMEG_2194|M.smegmatis_MC2_155 PDSEVAMDAAEQHRLHVNRWFYDCPPDFHRKLGDMYVSDPAYVARYDESF
TH_2460|M.thermoresistible__bu --------------------------------------------------
MAV_3222|M.avium_104 --------------------------------------------------
MAB_4723c|M.abscessus_ATCC_199 RTRADLAERMSLHAKRLYADHPGMLNSTADAPRGQRFAESTVARALLDLF
Mb3367|M.bovis_AF2122/97 --------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------
MMAR_0009|M.marinum_M GLPGLAAYCREAIHANANRTGG----
MUL_0011|M.ulcerans_Agy99 GLPGLAAYCREAIHANANRAGG----
Mvan_1919|M.vanbaalenii_PYR-1 GLPGLAAYCREAIHANADRAGQ----
Mflv_0148|M.gilvum_PYR-GCK GLPGLARYCRDAIHANADRS------
MSMEG_2194|M.smegmatis_MC2_155 GLPGLAAFCRDAIHANADRAGGDVTG
TH_2460|M.thermoresistible__bu --------------------------
MAV_3222|M.avium_104 --------------------------
MAB_4723c|M.abscessus_ATCC_199 NPAQLDVLIRVGAAVRSEPGEG----