For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVPGTDQDLLQIGEVAARTELSVKTIRHYDEVGLVVPSARSAGGFRLYTTDDVRRLMAIRRMKPLGFTLD EMGRLLDALAVLDKPAATAAERATAAEVVSDCHTRAQEACDRLTRHLAYARELTEQLAKVRG*
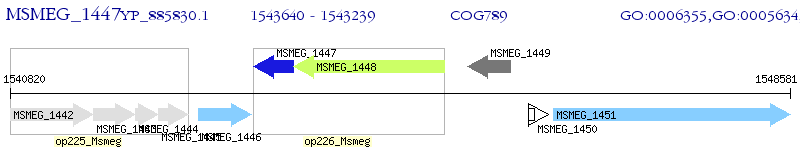
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1447 | - | - | 100% (133) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_2194 | - | 2e-13 | 48.53% (68) | MerR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3180 | - | 2e-08 | 38.20% (89) | transcriptional regulator, MerR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3367 | - | 1e-09 | 40.74% (81) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0148 | - | 1e-12 | 41.10% (73) | MerR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3334 | - | 1e-09 | 40.74% (81) | MerR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2309c | - | 9e-09 | 34.12% (85) | putative transcriptional regulator |
| M. marinum M | MMAR_0009 | - | 2e-14 | 47.14% (70) | transcriptional regulator |
| M. avium 104 | MAV_3222 | - | 6e-10 | 38.61% (101) | transcriptional regulator, MerR family protein |
| M. thermoresistible (build 8) | TH_0931 | soxR | 9e-05 | 36.23% (69) | redox-sensitive transcriptional activator SoxR |
| M. ulcerans Agy99 | MUL_0011 | - | 2e-14 | 47.14% (70) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1160 | - | 8e-44 | 72.00% (125) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1447|M.smegmatis_MC2_155 MTVPGTDQDLLQIGEVAARTELSVKTIRHYDEVGLVVPSARSAGGFRLYT
Mvan_1160|M.vanbaalenii_PYR-1 ------MAELLQIGEVAARTELSVKTIRHYDEVGLVTPSSRSGGGFRLYT
Mb3367|M.bovis_AF2122/97 ----------MKISEVAALTNTSTKTLRFYENSGLLPPPARTASGYRNYG
Rv3334|M.tuberculosis_H37Rv ----------MKISEVAALTNTSTKTLRFYENSGLLPPPARTASGYRNYG
MMAR_0009|M.marinum_M -------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRAASGYRVYT
MUL_0011|M.ulcerans_Agy99 -------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRAASGYRVYT
Mflv_0148|M.gilvum_PYR-GCK -MTQETRMEELTVGEVAKRFGITIRTLHHYDHIGLLTPEQRNASGYRVYT
MAB_2309c|M.abscessus_ATCC_199 ---MKGKTELLPIGEVARRSGIAVSAVRYYADIGLVP-AERTSGNVRVFR
TH_0931|M.thermoresistible__bu ----MGAHELTPS-EMSARSGVAVSALHFYEREGLIT-SRRTAGNQRRYP
MAV_3222|M.avium_104 ---------MLTIGEVARRSGVAATTLRYYEQIGLLPAPTRLG-GQRRYD
::: : :::.* **: * . . * :
MSMEG_1447|M.smegmatis_MC2_155 TDDVRRLMAIRRMKPLGFTLDEMGRLLD----------------------
Mvan_1160|M.vanbaalenii_PYR-1 DDDVSRLIAIRRMKPLGFTLDEMRGLLD----------------------
Mb3367|M.bovis_AF2122/97 PEIVDRLRFIHRGQAAGLALQEVRQILA----------------------
Rv3334|M.tuberculosis_H37Rv PEIVDRLRFIHRGQAAGLALQEVRQILA----------------------
MMAR_0009|M.marinum_M SADLTRLSQVIVYRRLELPLDEIARLLDEGNAVSHLVRQRERVMARLDEM
MUL_0011|M.ulcerans_Agy99 SADLTRLSQVIVYRRLELPLDEIARLLDEGNAVSHLVRQRERVMARLDEM
Mflv_0148|M.gilvum_PYR-GCK SADLTRLSQIIVYRRLEMSLDEIASVLDEGDEISHLRRQRERVMSRLDEL
MAB_2309c|M.abscessus_ATCC_199 RHALRRISLIRVATGFGIPLSEVAEVLS----------------------
TH_0931|M.thermoresistible__bu REMLRRVAFIRMSQRLGIPLARIREALA----------------------
MAV_3222|M.avium_104 EAVLSRLEVIALCKTAGFALDEIQRLFAD---------------------
: *: : :.* .: :
MSMEG_1447|M.smegmatis_MC2_155 -------------ALAVLDKPAATA---------AERATAAEVVSDC---
Mvan_1160|M.vanbaalenii_PYR-1 -------------ALDTLASPDTGA---------QARAEASAYLAVC---
Mb3367|M.bovis_AF2122/97 -------------IHDRGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHL
Rv3334|M.tuberculosis_H37Rv -------------IHDRGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHL
MMAR_0009|M.marinum_M KDLVEAIDQALEKAMTNTPMTDDDMRELFGDGFDDYRAETEQKWGETTEW
MUL_0011|M.ulcerans_Agy99 KDLVEAIDLALEKAMTNTPMTDDDMRELFGDGFDDYRVETEQKWGETAEW
Mflv_0148|M.gilvum_PYR-GCK TELVGAIDQALENAMTNTPMTDDDMRELFGDSFDDYQAEAEQKWGETAEW
MAB_2309c|M.abscessus_ATCC_199 -------------TLPDYRAPSTRDWQRIS---RRWHAHLEARKNAIADM
TH_0931|M.thermoresistible__bu -------------TLPSDRVPTSKDWAELS---AGWRADLDERILHLQRL
MAV_3222|M.avium_104 ---------------DAPGRPASRALAR------AKLAQIDAQLDSLTRA
. .
MSMEG_1447|M.smegmatis_MC2_155 ---HTRAQ-EACDRLTRHLAYARELTEQLAKVRG----------------
Mvan_1160|M.vanbaalenii_PYR-1 ---HGRAQ-EACVTLTRQLGYAREFMEQLAAASGEDVTPH----------
Mb3367|M.bovis_AF2122/97 QTLLDHAS-YGPPTEHDHSTVCWILESDLDEPTAIEVSDIHA--------
Rv3334|M.tuberculosis_H37Rv QTLLDHAS-YGPPTEHDHSTVCWILESDLDEPTAIEVSDIHA--------
MMAR_0009|M.marinum_M KESQRRTRSYGKDEWVRIKAEGQAVEKTLSDAFRAGLPADSEEAMNAAEQ
MUL_0011|M.ulcerans_Agy99 KESQRRTKSYGKDEWVRIKAEGQAIEKALSDAFRAGLPADSEEAMNAAEQ
Mflv_0148|M.gilvum_PYR-GCK KESQRRATSYGKDDWVRIKAEGQAVEKALADAFRAGLPADSEEAMNAAEQ
MAB_2309c|M.abscessus_ATCC_199 QEKLTGCIGCGCLSMTKCALFNPADILADDGSTGALLLADLG--------
TH_0931|M.thermoresistible__bu RDNLADCIGCGCLSLKTCALSNPDDALAARGPGAARL-------------
MAV_3222|M.avium_104 RAVIEWGMRCTCPSIDACTCGIHPTRPAPVGPDVRSARRF----------
MSMEG_1447|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1160|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3367|M.bovis_AF2122/97 --------------------------------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0009|M.marinum_M HRRHVNRWFYDCPPAFHRSLGDMYVSDPRYIATYDESFGLPGLAAYCREA
MUL_0011|M.ulcerans_Agy99 HRRHVNRWFYDCPPAFHRNLGDMYVSDPRYIATYDESFGLPGLAAYCREA
Mflv_0148|M.gilvum_PYR-GCK HRHHVNRWFYDCPPAFHRTLGDMYVSDPRYVATYDESFGLPGLARYCRDA
MAB_2309c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0931|M.thermoresistible__bu --------------------------------------------------
MAV_3222|M.avium_104 --------------------------------------------------
MSMEG_1447|M.smegmatis_MC2_155 ----------
Mvan_1160|M.vanbaalenii_PYR-1 ----------
Mb3367|M.bovis_AF2122/97 ----------
Rv3334|M.tuberculosis_H37Rv ----------
MMAR_0009|M.marinum_M IHANANRTGG
MUL_0011|M.ulcerans_Agy99 IHANANRAGG
Mflv_0148|M.gilvum_PYR-GCK IHANADRS--
MAB_2309c|M.abscessus_ATCC_199 ----------
TH_0931|M.thermoresistible__bu ----------
MAV_3222|M.avium_104 ----------