For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDERYRQSFQLEPVTRIADRVADQVRDYIVANDLEPGARLPSERRLAELVGSSRPTVSQALRSLSVTGLI EIRPGAGAFVLRKPTTAVGTGLQMMIELEPDSIGEAVDMRHLLEMAAVERITAKPTLELDSIEEALERLR AAKGSAAEWIAADTNFHVTFIRLAENRFLTSVFESVHDAVVARAYQSWIVANEPPPWLAGKEFAAQIALH EPIVAALRANDGAQLVAALTRHQKALEAHMRLHG
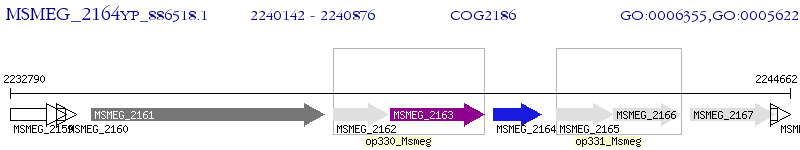
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2164 | - | - | 100% (244) | transcriptional regulator, putative |
| M. smegmatis MC2 155 | MSMEG_2173 | - | 4e-21 | 29.20% (226) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2605 | - | 6e-15 | 27.88% (165) | transcriptional regulator, GntR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 3e-13 | 29.11% (237) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0016 | - | 4e-14 | 28.40% (169) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0494 | - | 3e-13 | 29.11% (237) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0068 | - | 6e-13 | 27.31% (216) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0820 | - | 5e-12 | 31.05% (190) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2560 | - | 3e-13 | 29.82% (171) | regulatory protein GntR, HTH |
| M. thermoresistible (build 8) | TH_4408 | - | 8e-13 | 26.99% (226) | PUTATIVE regulatory protein GntR, HTH |
| M. ulcerans Agy99 | MUL_4564 | - | 6e-12 | 31.22% (189) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0952 | - | 4e-13 | 28.07% (171) | GntR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0505|M.bovis_AF2122/97 MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
Rv0494|M.tuberculosis_H37Rv MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
MMAR_0820|M.marinum_M ----MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAE
MUL_4564|M.ulcerans_Agy99 ----MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAE
Mflv_0016|M.gilvum_PYR-GCK MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVE
Mvan_0952|M.vanbaalenii_PYR-1 --------MPLSTARRTGLVDQVIEQLRRAVAGGEWRVGERIPNETVLVE
MAV_2560|M.avium_104 --------MEVTSLREPKMADRVATVLRRMFIRGEITEGTMLPPESELME
TH_4408|M.thermoresistible__bu ----VEDYMEVTNLREPKMADRVATVLRRMFIRGEIPEGTMLPPESELME
MSMEG_2164|M.smegmatis_MC2_155 --MDERYRQSFQLEPVTRIADRVADQVRDYIVANDLEPGARLPSERRLAE
MAB_0068|M.abscessus_ATCC_1997 ----------MIRTVAISASEEAYRSVKESILNGDIP-GGELLSEGEISA
. :. : . * : * :
Mb0505|M.bovis_AF2122/97 RLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPEGLTHPAVVEALVRK
Rv0494|M.tuberculosis_H37Rv RLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPEGLTHPAVVEALVRK
MMAR_0820|M.marinum_M QLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQSLTHPAVVEALVRK
MUL_4564|M.ulcerans_Agy99 QLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQSLTHPAVVEALVRK
Mflv_0016|M.gilvum_PYR-GCK KFGVGRNTVREAVRALAHAGILEVRQGDGTYVR-----ATSEVSGALRRL
Mvan_0952|M.vanbaalenii_PYR-1 SLGVGRNTVREAVRALAHAGILEVRQGDGTYVR-----ATSEVSGALRRL
MAV_2560|M.avium_104 RFGVSRPTLREAFRVLESESLIQVQRGVRGGAR-----VTRPRRETLARY
TH_4408|M.thermoresistible__bu RFGVSRPTLREAFRVLESESLIVIQRGVRGGAR-----VTRPRRETLARY
MSMEG_2164|M.smegmatis_MC2_155 LVGSSRPTVSQALRSLSVTGLIEIRPGAGAFVLR---KPTTAVGTGLQMM
MAB_0068|M.abscessus_ATCC_1997 RMGCSRTPVREAFLRLETEGWLRLYPKRGALVVP----ITPEER------
.* .* .: :.. * . : .
Mb0505|M.bovis_AF2122/97 LG------PDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQ
Rv0494|M.tuberculosis_H37Rv LG------PDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQ
MMAR_0820|M.marinum_M LG------PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAEVA
MUL_4564|M.ulcerans_Agy99 LG------PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAEVA
Mflv_0016|M.gilvum_PYR-GCK CG-------EELREVLEVRRALEVEAARLAASRRTPADVAEMRGLLARRD
Mvan_0952|M.vanbaalenii_PYR-1 CG-------AELREVLQVRRALEVEGARLAAAHCTPADIAEMRTLLARRD
MAV_2560|M.avium_104 AGLILEYEGVTVKDVYDARVTLEVPMVEQLAKDRNPKVIAELEQIVERES
TH_4408|M.thermoresistible__bu AGLILEYEEVTLKDVYEARMTIEMPMVVELAKKRDPAVIAELEAIVANES
MSMEG_2164|M.smegmatis_MC2_155 IELE----PDSIGEAVDMRHLLEMAAVERITAKPT-LELDSIEEALERLR
MAB_0068|M.abscessus_ATCC_1997 ------------RHVVDARRVIEGAAAARIAERGAPQSLLSDLSALIEVQ
. : * : : :
Mb0505|M.bovis_AF2122/97 QADTA----AARQAADLAYFRVLIHSTRN---RALGLLYRWVEHAFGGRE
Rv0494|M.tuberculosis_H37Rv QADTA----AARQAADLAYFRVLIHSTRN---RALGLLYRWVEHAFGGRE
MMAR_0820|M.marinum_M DADTA----PARQAADLAYFRVLIHGTRN---RALGLLYRWVEQAFGGRE
MUL_4564|M.ulcerans_Agy99 DADTA----PARQAADLAYFRVLIHGTRN---RALGLLYRWVEQAFGGRE
Mflv_0016|M.gilvum_PYR-GCK ERDSAG-DVEAFARIDTEFHLAVVRCARN---NTLIELYRGLLEAVTASV
Mvan_0952|M.vanbaalenii_PYR-1 ECAATD-DTDAFARTDAEFHLAVVRGSGN---RTLTELYRGLIEVVTASV
MAV_2560|M.avium_104 QLNPG----SEAVDQLTDFHAAIARLSGNSTLQIVSDMLHHIIEKANRSL
TH_4408|M.thermoresistible__bu NLKPG----SETVDQLTDFHAAVARLSGNKTLQIVADMLHHIVKTANRSL
MSMEG_2164|M.smegmatis_MC2_155 AAKGS---AAEWIAADTNFHVTFIRLAEN---RFLTSVFESVHDAVVARA
MAB_0068|M.abscessus_ATCC_1997 LGRAAQGDAAGFAAADVDFHRAIVAKLGN---PLLENFYDSLRERQRRMA
:. .. * : . : .
Mb0505|M.bovis_AF2122/97 HALTG-----AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASAL
Rv0494|M.tuberculosis_H37Rv HALTG-----AYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASAL
MMAR_0820|M.marinum_M HELTG-----AYEDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASAL
MUL_4564|M.ulcerans_Agy99 HDLTG-----AYEDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASAL
Mflv_0016|M.gilvum_PYR-GCK AHTR---------HVPVDVCDHGVLLDHIAAGDPDLAARTAGEFLDR---
Mvan_0952|M.vanbaalenii_PYR-1 ATTR---------HVPVEMVDHGVLVDQIAAGDAEQAARTAGELLDR---
MAV_2560|M.avium_104 QPTKGTR---AEQAVRRSAKTHRMVLDMIKEGDAEKAGELWKRHLQKAEE
TH_4408|M.thermoresistible__bu QPTEGTR---AEQAVRRSAKTHRMVLELIKAGDAEKAGELWQRHLQKAEE
MSMEG_2164|M.smegmatis_MC2_155 YQSWIVANEPPPWLAGKEFAAQIALHEPIVAALRANDGAQLVAALTRHQK
MAB_0068|M.abscessus_ATCC_1997 AAAIG----VDPVRVARSVAGHRALVDALAAGDAARFSTELVEHMKVVTE
. : : . . :
Mb0505|M.bovis_AF2122/97 RMVKSYRDRA-------
Rv0494|M.tuberculosis_H37Rv RMVKSYRDRA-------
MMAR_0820|M.marinum_M RMIMCYRAAYAGEDPER
MUL_4564|M.ulcerans_Agy99 RMIMCYRAAYAGEDPER
Mflv_0016|M.gilvum_PYR-GCK -IINGLGSADSS-----
Mvan_0952|M.vanbaalenii_PYR-1 -ILAGVDSADSC-----
MAV_2560|M.avium_104 FVLTGAELSTVVDLLE-
TH_4408|M.thermoresistible__bu FVLSGAEMSTVVDLLE-
MSMEG_2164|M.smegmatis_MC2_155 ALEAHMRLHG-------
MAB_0068|M.abscessus_ATCC_1997 VD---------------