For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARGADFLQLDPASAPARGLTSWLVDALRSAIADGRLPPGVRLPATRTLAGELAVSRGVVVEAYRRLADE GLVSGRTGGGTRVLARPVRPPRRARTAAPLDRPRLPRSRLPTGEGIDLSPGVPDLSAFPRSTWLRAERAV LSETAPEDLGYGDPRGHPRLRAALAPWLGRTRGLRVDPDDILVVAGVAQAVALLAQQLRREGLDTIAVED PGSRGAVDELEYWGLRAMPVPVDDEGIQVQSLGETGAAAVFLTPAHQFPTGVVLGPRRRRELLAWDGELV IEDDYDAEYRYDRAPVPALHPSAPERIAYAGSTSKTLAPALRLGWLVAPRRRHPDLVAAKHATDLGSPTV PQLVLARLLESGEYDRHIRLVRARHRARRDALLETLATALPAARVTGVAAGLHLLVTLLADVDDVALADE LRRAGVLVHPLSWHRRTPGPPGLVLGYASHSPDRLREAGSIIARIAE
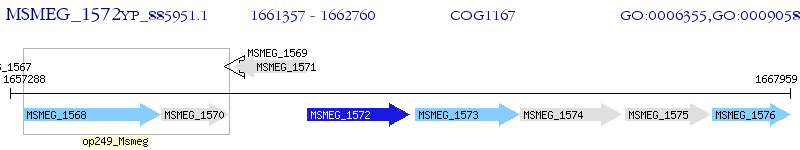
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1572 | - | - | 100% (467) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_4140 | - | 1e-84 | 42.48% (452) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5760 | - | 1e-83 | 42.08% (461) | GntR-family protein regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1211 | - | 8e-13 | 29.77% (346) | N-succinyldiaminopimelate aminotransferase |
| M. gilvum PYR-GCK | Mflv_1181 | - | 4e-77 | 41.47% (422) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1178 | - | 4e-13 | 29.57% (345) | N-succinyldiaminopimelate aminotransferase |
| M. leprae Br4923 | MLBr_01488 | - | 4e-09 | 27.45% (306) | N-succinyldiaminopimelate aminotransferase |
| M. abscessus ATCC 19977 | MAB_0196c | - | 2e-75 | 39.83% (467) | GntR family transcriptional regulator |
| M. marinum M | MMAR_2376 | - | 2e-22 | 33.43% (329) | GntR family transcriptional regulator |
| M. avium 104 | MAV_3214 | - | 7e-22 | 29.76% (457) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1831 | - | 1e-13 | 29.50% (322) | PROBABLE AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_1552 | - | 2e-22 | 33.43% (329) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5625 | - | 1e-75 | 40.62% (453) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1211|M.bovis_AF2122/97 --------------------------------------------------
Rv1178|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01488|M.leprae_Br4923 --------------------------------------------------
TH_1831|M.thermoresistible__bu --------------------------------------------------
Mflv_1181|M.gilvum_PYR-GCK MVSSWSNSG-------SRDLHLDLREALRPGARGNRNALVAALREAVRSG
Mvan_5625|M.vanbaalenii_PYR-1 -MTSWANSA-------TRDLHLDLRDALVPGARGTKDALISALRDAARSG
MAB_0196c|M.abscessus_ATCC_199 -MSSWANSAKAESRPLSHDLHLDLRQSITPGTRGVRELLTSALRDAVRSG
MSMEG_1572|M.smegmatis_MC2_155 -MARGAD-------------FLQLDPASAP-ARGLTSWLVDALRSAIADG
MMAR_2376|M.marinum_M ------------------------MPVQYRIAGGGAESIAASIESAISQG
MUL_1552|M.ulcerans_Agy99 ------------------------MPVQNRIAGGGAGSIAASIESAISQG
MAV_3214|M.avium_104 ------------------------MAVQKSITGTGAESIAASIEAAISAG
Mb1211|M.bovis_AF2122/97 -------------------MSASLPVFPWDTLADAKALAGAHPDG-----
Rv1178|M.tuberculosis_H37Rv -------------------MSASLPVFPWDTLADAKALAGAHPDG-----
MLBr_01488|M.leprae_Br4923 -------------MSPFGRVSATLPVFPWDTLADAKAVAESHPDG-----
TH_1831|M.thermoresistible__bu --------------------------------------------------
Mflv_1181|M.gilvum_PYR-GCK RLSAGTTLPPSRALAADLGLARNTVADAYAELVAEGWLASRQGAGTWVVD
Mvan_5625|M.vanbaalenii_PYR-1 RLTAGTMLPSSRTLATDLGLARNTVAEAYAELVAEGWLASRQGAGTWVAD
MAB_0196c|M.abscessus_ATCC_199 RLTVGSMLPPSRTLASDLGLARNTVAEAYAELVAEGWLESRQGSGTRVVN
MSMEG_1572|M.smegmatis_MC2_155 RLPPGVRLPATRTLAGELAVSRGVVVEAYRRLADEGLVSGRTGGGTRVLA
MMAR_2376|M.marinum_M GLAPGDALPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRD
MUL_1552|M.ulcerans_Agy99 GLAPGDALPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRD
MAV_3214|M.avium_104 SLAPGDALPPVRELAARLGVNANTAAAAYRLLRHRGAVETAGRRGTRVRH
Mb1211|M.bovis_AF2122/97 -----------------------------IVDLSVGTPVDPVAP---LIQ
Rv1178|M.tuberculosis_H37Rv -----------------------------IVDLSVGTPVDPVAP---LIQ
MLBr_01488|M.leprae_Br4923 -----------------------------IVDLSVGTPVDPVAP---LIQ
TH_1831|M.thermoresistible__bu -----------------------------------------VAP---VIR
Mflv_1181|M.gilvum_PYR-GCK TARASSQPPPVPP---RPHGARVAPVH----NLMPGSPDVAEFPRSQWAA
Mvan_5625|M.vanbaalenii_PYR-1 VT----TPAHTPP---RPHGVRVAPTH----NLMPGSPDVAEFPRSQWLA
MAB_0196c|M.abscessus_ATCC_199 RG-----KDQLPP---RPRGAGTPPPH----NLMPGSGDVTAFPRADWLA
MSMEG_1572|M.smegmatis_MC2_155 RPVRPPRRARTAAPLDRPRLPRSRLPTGEGIDLSPGVPDLSAFPRSTWLR
MMAR_2376|M.marinum_M RP---ASTPRSSLDLGVPAGAR---------DLSTGNPAPELLP-----L
MUL_1552|M.ulcerans_Agy99 RP---ASTPRSSLDLGVPAGAR---------DLSTGNPAPELLP-----L
MAV_3214|M.avium_104 RP---ATTPRSLLGLDVPAGVR---------DLSTGNPHPALLP-----L
*
Mb1211|M.bovis_AF2122/97 EALAAASAAPGYPATAGTARLRESVVAALARRYGITR---LTEAAVLPVI
Rv1178|M.tuberculosis_H37Rv EALAAASAAPGYPATAGTARLRESVVAALARRYGITR---LTEAAVLPVI
MLBr_01488|M.leprae_Br4923 EALAAASSVAGYPTTAGTARLREAAVAALDRRYGITG---LTEAAVLPVI
TH_1831|M.thermoresistible__bu AALAEASAAPGYPTTAGTPALRASAAAALERRYGITG---LTPDAVLPAI
Mflv_1181|M.gilvum_PYR-GCK SARRALTHAPTDAFRMGDPRGRPELRSALAEYLARARGVRTSPESLVICA
Mvan_5625|M.vanbaalenii_PYR-1 STRRALGGAPTEALRMGDPRGRPELRSALAEYLARARGVRTSPESIVICA
MAB_0196c|M.abscessus_ATCC_199 SARRALNHAPHEALRVGDPRGRHELRSAVAEYVSRVRGVRTTPDSILICA
MSMEG_1572|M.smegmatis_MC2_155 AERAVLSETAPEDLGYGDPRGHPRLRAALAPWLGRTRGLRVDPDDILVVA
MMAR_2376|M.marinum_M AAVG-WPDGSVLPLLYGAPAMSAELVDYARAALAADG---VAADHLAVTN
MUL_1552|M.ulcerans_Agy99 AAVG-WPDGSVLPLLYGAPAMSAELVDYARAALAADG---VAADHLAVTN
MAV_3214|M.avium_104 AAAAPARPASGRAVLYGEPAISPALAEYARAALSADG---VPADHLALTS
. * . . :
Mb1211|M.bovis_AF2122/97 GTKELIAWLPTLLGLGGADLVVVPELAYPTYDVGARLAGTRVLRADA---
Rv1178|M.tuberculosis_H37Rv GTKELIAWLPTLLGLGGADLVVVPELAYPTYDVGARLAGTRVLRADA---
MLBr_01488|M.leprae_Br4923 GTKELIAWLPTLLGLRAGDVVVVPELAYPTYNVGARLAGARVLRADS---
TH_1831|M.thermoresistible__bu GTKELIAWLPTLLGLGGDDTVVVPELAYPTYDVGARLAGARVFRADS---
Mflv_1181|M.gilvum_PYR-GCK GVRHAVELLARTLG----GPVAVEAYGLFVFRDAIAACGVQTVPIGVDDD
Mvan_5625|M.vanbaalenii_PYR-1 GVRHAVELLTRALG----GPIAVESYGLFVFRDAIAAAGASTVPIGVDDR
MAB_0196c|M.abscessus_ATCC_199 GTRHAVEILTRMYGLA--RPIAVEAYGLFIFRDCIEATGGSSTPIGFDGE
MSMEG_1572|M.smegmatis_MC2_155 GVAQAVALLAQQLRREGLDTIAVEDPGSRGAVDELEYWGLRAMPVPVDDE
MMAR_2376|M.marinum_M GALDGIERALSANLRPG-DRVAVEDPGWANLLDLLAALGLPVEPMGVDDD
MUL_1552|M.ulcerans_Agy99 GALDGIERALSANLRPG-DRVAVEDPGWANLLDLLAALGLPVEPMGVDDD
MAV_3214|M.avium_104 GALDGIERALTAHLRPG-DRVAVEDPGWANLLDLLAALGFSAEPVRVDDA
*. . : :.* . *
Mb1211|M.bovis_AF2122/97 ---LTQLG----PQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARGRGVL
Rv1178|M.tuberculosis_H37Rv ---LTQLG----PQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARGRGVL
MLBr_01488|M.leprae_Br4923 ---LTQLG----LQSPALFYLNSPSNPTGQVLAVNHLRKMVGWARNRGVV
TH_1831|M.thermoresistible__bu ---LTQLG----PQTPALIYLNSPSNPTGRVLGVDHLRKVVGWARDRGVI
Mflv_1181|M.gilvum_PYR-GCK GAVIADLD---DTSARSVLLTPAHHNPLGMPLHPSRRAAVIEWATRTGGY
Mvan_5625|M.vanbaalenii_PYR-1 GAVVDELD---GLDVPAVLLTPAHHNPLGMPLHSSRRTAVIEWAQRTGGF
MAB_0196c|M.abscessus_ATCC_199 GAVVSDLE---NLDTSAALITPAHHFPHGLPLHPARRTAVVDWARRTDSY
MSMEG_1572|M.smegmatis_MC2_155 GIQVQSLG---ETGAAAVFLTPAHQFPTGVVLGPRRRRELLAWD---GEL
MMAR_2376|M.marinum_M GPLVADLQRALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEE
MUL_1552|M.ulcerans_Agy99 GPLVADLQRALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEE
MAV_3214|M.avium_104 GPLPEDLARALGRGARALVLTTRAQNPTGAALSAERAAALRDLLSGRADD
.* : . * * * : :
Mb1211|M.bovis_AF2122/97 VVSDECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAG
Rv1178|M.tuberculosis_H37Rv VVSDECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAG
MLBr_01488|M.leprae_Br4923 VVSDECYLGLGWDAEPFSVLHPLVCDGNHTGLLAVHSLSKSSSLAGYRAG
TH_1831|M.thermoresistible__bu VASDECYLGLGWDETPVSVLHPTVCGGDHTNLLAIHSLSKTSSLAGYRAG
Mflv_1181|M.gilvum_PYR-GCK ILDDDYDGEFRYDRQPVGALQALRP-----DRVAYLGSTSKSLSQALRLG
Mvan_5625|M.vanbaalenii_PYR-1 VLDDDYDGEFRYDRQPVGALQSLRP-----DRVAYLGSTSKSLSQTLRLG
MAB_0196c|M.abscessus_ATCC_199 VLEDDYDGEFRYDRQPVGALQSLDP-----DRVVYLGSASKSLAPALRLG
MSMEG_1572|M.smegmatis_MC2_155 VIEDDYDAEYRYDRAPVPALHPSAP-----ERIAYAGSTSKTLAPALRLG
MMAR_2376|M.marinum_M VLLVEDDHCAGIAGVPLHTLAGAT------QHWAFVRSAAKAYGPDLRLA
MUL_1552|M.ulcerans_Agy99 VLLVEDDHCAGIAGVPLHTLAGAT------QHWAFVRSAAKAYGPDLRLA
MAV_3214|M.avium_104 VLLVEDDHCAGISGAALHPVAGCT------THWAFVRSASKAYGPDLRVA
: : .. : . : .: * .
Mb1211|M.bovis_AF2122/97 FVVGDLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHE---RQQRER
Rv1178|M.tuberculosis_H37Rv FVVGDLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHE---RQQRER
MLBr_01488|M.leprae_Br4923 FVAGDPDLVAELLAVRKHAGMMVPTPVQAAMVAALGDDAHE---REQRDR
TH_1831|M.thermoresistible__bu FVAGDPALVAELLAVRKHAGMMVPTPVQAAMVAALDDDAHE---RQQRER
Mflv_1181|M.gilvum_PYR-GCK WMALPAELVDQVVETAGGQQFHVDVVSQLTLADFLTAGHYDRHIRRMRMR
Mvan_5625|M.vanbaalenii_PYR-1 WMALPDNLIDAVLAAAGGQQFYVDAINQLTLADFIAGGHYDRHIRRMRMR
MAB_0196c|M.abscessus_ATCC_199 WMVLPPEQMDAAVAATGGAQWYVGAISQLTMADFIAGGHYDRHIRRMRGR
MSMEG_1572|M.smegmatis_MC2_155 WLVAPRRRHPDLVAAKHATDLGSPTVPQLVLARLLESGEYDRHIRLVRAR
MMAR_2376|M.marinum_M VLAGDQRTVERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQER
MUL_1552|M.ulcerans_Agy99 VLAGDQRTVERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQER
MAV_3214|M.avium_104 VLAGDQRTVDRVHGRLRLGPGWVSHVLQQLAVELWSDTAASELVATAERR
:. * . . *
Mb1211|M.bovis_AF2122/97 YAQRRAALLPALGSAGFAVDYS--DAGLYLWATRGEPCRDSVAWLAQR--
Rv1178|M.tuberculosis_H37Rv YAQRRAALLPALGSAGFAVDYS--DAGLYLWATRGEPCRDSAAWLAQR--
MLBr_01488|M.leprae_Br4923 YARRRDVLLPAVVSAGFTVDHS--EAGLYLWASRGEPCRDSVEWFAQR--
TH_1831|M.thermoresistible__bu YQRRRRELLPALQQAGFTVDHS--EAGLYLWATRGEPCRATVDWLARR--
Mflv_1181|M.gilvum_PYR-GCK YRRRRDMLVSALSP--FAVGIGGLAAGVNLLVTLPDGAEPELIRRAGE-A
Mvan_5625|M.vanbaalenii_PYR-1 YRRRRDVLVSALSP--FDVGIGGLAAGVNLLVTLPDGAEPEVMRRAGE-S
MAB_0196c|M.abscessus_ATCC_199 YRRRRDLLVQRLAG--YNVGVRGLSAGLHLLLTLPEGTESDVLYRAGE-A
MSMEG_1572|M.smegmatis_MC2_155 HRARRDALLETLATALPAARVTGVAAGLHLLVTLLADVDDVALADELRRA
MMAR_2376|M.marinum_M YADARGRLCAALSARGVAAHGR---SGLNVWIPVPD--ETAAITRLLN-R
MUL_1552|M.ulcerans_Agy99 YADARGRLCAALSARGVAAHGR---SGLNVWIPVPD--ETAAITRLLN-R
MAV_3214|M.avium_104 YADRRRRLCGALAERGVDAHGR---SGLNVWVRVPD--EAVAVSRLLG-A
: * * : . :*: :
Mb1211|M.bovis_AF2122/97 GILVAPGDFYGPGGAQH-------VRVALTAT-DERVAAAVGRLTC----
Rv1178|M.tuberculosis_H37Rv GILVAPGDFYGPGGAQH-------VRVALTAT-DERVAAAVGRLTC----
MLBr_01488|M.leprae_Br4923 GILVALGEFYGPGGCQH-------VRVALTAS-DERIAAVVARLS-----
TH_1831|M.thermoresistible__bu GILVAPGEFYGPRGAEH-------VRIALTAP-DERITAAAARLAQAG--
Mflv_1181|M.gilvum_PYR-GCK GIALVGLSMMRHPRAGSGVAERDGLIVGFAAAPEHAFPAAVSALCEVLRA
Mvan_5625|M.vanbaalenii_PYR-1 GLAVSGLSIMRHPMAGP--ADVDGLIVGFAAPPEHAFSAAVAALCDVLRA
MAB_0196c|M.abscessus_ATCC_199 GVALAGLARLRHPLAGPHIPNPDGVVVSFGTPADHGFGPAVDALCRVLAA
MSMEG_1572|M.smegmatis_MC2_155 GVLVHPLSWHRRTPGPP------GLVLGYASHSPDRLREAGSIIARIAE-
MMAR_2376|M.marinum_M GWAAAPGARFRIRSAPG-------IRVTIAGLSAEEIEPLADAIAEAIQR
MUL_1552|M.ulcerans_Agy99 GWAAAPGARFRVRSAPG-------IRVTIAGLSAEEIEPLADAIAGAIQR
MAV_3214|M.avium_104 GWAAAPGARFRMQTPAG-------IRITIADLTPDEIEPLADAVAQAVRP
* : : . . :
Mb1211|M.bovis_AF2122/97 ------
Rv1178|M.tuberculosis_H37Rv ------
MLBr_01488|M.leprae_Br4923 ------
TH_1831|M.thermoresistible__bu ------
Mflv_1181|M.gilvum_PYR-GCK SGL---
Mvan_5625|M.vanbaalenii_PYR-1 SGL---
MAB_0196c|M.abscessus_ATCC_199 TGL---
MSMEG_1572|M.smegmatis_MC2_155 ------
MMAR_2376|M.marinum_M TGRPTV
MUL_1552|M.ulcerans_Agy99 TGRPTV
MAV_3214|M.avium_104 SGRPIV