For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSLVSYALNGNVATVTLDDGKANALSPDMQAAINDALDQASLAAGSGEVKALVVAGNSKVFSGGFDLSVF ASGDAAAALGMLTGGFELAVRCLTFPVPVIMAATGPAIAMGSFLLLSGDHRVGSPRSRCQANEVAIGMTL PIAAIEIMRMRLTRAAFQRAISMAAVFSGDAAISAGWLDEIVEPDEVLDRAQQVATEAAALHTNAHVASK LKARADAVTAIRAGIDGLAAEFGS*
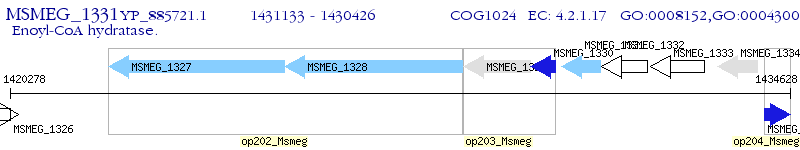
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1331 | - | - | 100% (235) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5639 | - | 3e-10 | 29.65% (199) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_0490 | - | 3e-10 | 26.54% (211) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0649c | echA3 | 2e-70 | 55.98% (234) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_5125 | - | 6e-88 | 71.79% (234) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0632c | echA3 | 2e-70 | 55.98% (234) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02118 | echA6 | 2e-08 | 28.73% (181) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1069c | - | 4e-11 | 28.09% (235) | enoyl-CoA hydratase |
| M. marinum M | MMAR_0962 | echA3 | 8e-67 | 54.27% (234) | enoyl-CoA hydratase, EchA3 |
| M. avium 104 | MAV_4534 | - | 5e-69 | 55.56% (234) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_1874 | echA3 | 6e-87 | 70.09% (234) | PROBABLE ENOYL-CoA HYDRATASE ECHA3 (ENOYL HYDRASE) |
| M. ulcerans Agy99 | MUL_0713 | echA3 | 6e-67 | 54.27% (234) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_1226 | - | 7e-87 | 70.94% (234) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0649c|M.bovis_AF2122/97 -MSDPVSYTRKDSIAVISMDDGK-VNALGPAMQQALNAAIDNADRDD---
Rv0632c|M.tuberculosis_H37Rv -MSDPVSYTRKDSIAVISMDDGK-VNALGPAMQQALNAAIDNADRDD---
MMAR_0962|M.marinum_M -MSGPVTYTHDDAIGVIRMDDGK-VNVLGPTMQQALNEAIDAADRDN---
MUL_0713|M.ulcerans_Agy99 -MSGPVTYTHDDAIGVIRMDDGK-VNVLGPTMQQALNEAIDAADRDN---
MAV_4534|M.avium_104 -MSGPVHYSTKGPVAVIRMDDGK-VNALGPTMQQALGEAIDRAEADN---
Mflv_5125|M.gilvum_PYR-GCK -MTGTVSYDLADSVATVTLDDGK-VNVLGPAMQAAINNALDRAEADK---
Mvan_1226|M.vanbaalenii_PYR-1 -MTSTVSYEFADSVATISLDDGK-VNVLGPAMQAAINEALDRAEADK---
TH_1874|M.thermoresistible__bu -MSSTVEYQLTDSVATITMDDGK-VNVLSPAMQGNLNEALDRAEQDG---
MSMEG_1331|M.smegmatis_MC2_155 -MSSLVSYALNGNVATVTLDDGK-ANALSPDMQAAINDALDQASLAAGSG
MAB_1069c|M.abscessus_ATCC_199 MSDTLVQYEVEGRAARLTLDSPHNRNALSSGLVRQLREGLQRAAADQT--
MLBr_02118|M.leprae_Br4923 ----MIGITQAEGVMTIELQRPERRNALNSQLIEKLREAVQKASSADSEV
: : :: . *.*.. : : .:: *
Mb0649c|M.bovis_AF2122/97 -VGALVITGNGRVFSGGFDLKILTSGEVQPAIDMLRGGFELAYR-LLSYP
Rv0632c|M.tuberculosis_H37Rv -VGALVITGNGRVFSGGFDLKILTSGEVQPAIDMLRGGFELAYR-LLSYP
MMAR_0962|M.marinum_M -VGALVIAGNHRVFSGGFDLKVLTSGEAKPAIDMLRGGFELSYR-LLSYP
MUL_0713|M.ulcerans_Agy99 -VGALVIAGNHRVFSGGFDLKVLTSGEAKPAIDMLRGGFELSYR-LLSYP
MAV_4534|M.avium_104 -AGALVITGNERVFSGGFDLKILTSGEVQPAVNMLRGGFELAYR-LLSYP
Mflv_5125|M.gilvum_PYR-GCK -AKAVVLAGNSRVFSGGFDLGVFQSGDAAAALGMLSGGFELSVR-TLTFP
Mvan_1226|M.vanbaalenii_PYR-1 -AKAVVLAGNSRVFSGGFDLSVFQSGDAAAALGMLAGGFELSLR-TLTFP
TH_1874|M.thermoresistible__bu -AKAVVLAGNSRVFSGGFDLSVFSSGDVDAALGMLRGGFELAVR-ALTFP
MSMEG_1331|M.smegmatis_MC2_155 EVKALVVAGNSKVFSGGFDLSVFASGDAAAALGMLTGGFELAVR-CLTFP
MAB_1069c|M.abscessus_ATCC_199 -VRAVVLTHTGNTFCAGADLSEASEGSPQDIAKDRAGQLTALLRDILELP
MLBr_02118|M.leprae_Br4923 STRVIVLTGQGTVFCAGADLSG------DAFAADYPDRLIELHRVMDAVP
. .:*:: .*..* ** . : * *
Mb0649c|M.bovis_AF2122/97 KPVVMACTGHAIAMGAFLLSCGDHRVAAHAYNIQANEVAIGMTIPYAALE
Rv0632c|M.tuberculosis_H37Rv KPVVMACTGHAIAMGAFLLSCGDHRVAAHAYNIQANEVAIGMTIPYAALE
MMAR_0962|M.marinum_M KPVVIACTGHAIAMGAFLLCSGDHRVAAHAYNVQANEVAIGMTIPYAAME
MUL_0713|M.ulcerans_Agy99 KPVVIACTGHAIAMGAFLLCSGDHRVAAHAYNIQANEVAIGMTIPYAAME
MAV_4534|M.avium_104 KPVVMACTGHAIAMGAFLLSSGDHRVAAHAYNIQANEVAIGMVIPYAAME
Mflv_5125|M.gilvum_PYR-GCK VPVIIAATGPAIAMGSFLLLSGDHRVGQPKTRCQAIEVAIGMTIPIAALE
Mvan_1226|M.vanbaalenii_PYR-1 APVIVAATGPAIAMGSFLLLSTDHRVGQPKSRCQAIEVAIGMTIPISALE
TH_1874|M.thermoresistible__bu VPVIMAATGPAIAMGSFLLLCGDHRVGSERTRCQANEVAIGMILPVAALE
MSMEG_1331|M.smegmatis_MC2_155 VPVIMAATGPAIAMGSFLLLSGDHRVGSPRSRCQANEVAIGMTLPIAAIE
MAB_1069c|M.abscessus_ATCC_199 LPVIIAVNGHVRAGGFGLVGAADIAVAGPDSTFALTEARIGVAPSIISLT
MLBr_02118|M.leprae_Br4923 MPVIGAINGPAIGAGLQLAMQCDLRVVAPDAYFQFPTSKYGLALDNWSIR
**: * .* . . * * * * *: ::
Mb0649c|M.bovis_AF2122/97 IMKLRLTRSAYQQATGLAKTFFGETALAAGFIDEIALPEVVVSRAEEAAR
Rv0632c|M.tuberculosis_H37Rv IMKLRLTRSAYQQATGLAKTFFGETALAAGFIDEIALPEVVVSRAEEAAR
MMAR_0962|M.marinum_M VLKLRLTPSAYQQAAGLAKTFFGETALAAGFIDEISLPEVVLSRAEEAAR
MUL_0713|M.ulcerans_Agy99 VLKLRLTPSAYQQAAGLAKTFFGETALAAGFIDEISLPEVVLSRAEEAAR
MAV_4534|M.avium_104 IMKLRLTPSAYQQASGLAKTFFGETALAAGFVDEIVLPEVVVDRAEEAAA
Mflv_5125|M.gilvum_PYR-GCK IMRARLTPAAFERSAALASTYVGDDAVAAGWLDEIVEADQVLTRAQQVAA
Mvan_1226|M.vanbaalenii_PYR-1 IMRMRLTPAAFERGAALAATFAGDDAVAGGWLDEIVPADQVLARAQQVAG
TH_1874|M.thermoresistible__bu IMRMRLTPAAYQHAVSLATTFTGDAAIRAGWLDEVVDTDRVLPRAREFAA
MSMEG_1331|M.smegmatis_MC2_155 IMRMRLTRAAFQRAISMAAVFSGDAAISAGWLDEIVEPDEVLDRAQQVAT
MAB_1069c|M.abscessus_ATCC_199 LLP-RLTSRAAGRYFVTGEKFDARVAAEIGLVTQSVDSAEAVEAAVTSLV
MLBr_02118|M.leprae_Br4923 RLSSLVGHGRARAMLLTAEKLTADIALQTGMANRIG----ALADAQAWAA
: : . . * * . .: *
Mb0649c|M.bovis_AF2122/97 EFAG-LNQHAHAATKLRSRADALTAIRAGIDGIAAEFGL-----------
Rv0632c|M.tuberculosis_H37Rv EFAG-LNQHAHAATKLRSRADALTAIRAGIDGIAAEFGL-----------
MMAR_0962|M.marinum_M EFAG-LNQQAHNATKLRARAEALKAIRAGIDGIEAEFGL-----------
MUL_0713|M.ulcerans_Agy99 EFAG-LNQQAHNATKLRARAEALKAIRAGIDGIEAEFGL-----------
MAV_4534|M.avium_104 EFAE-LNQRAHAATKLRARADALAAIRAGIDGIEAEFGL-----------
Mflv_5125|M.gilvum_PYR-GCK EAATTLHAGAHLATKLKARKYALDAIRAGIDGLATEFTLG----------
Mvan_1226|M.vanbaalenii_PYR-1 EAAATLHPGAHLATKLKARKPALDAIRAGIDGLATEFSLG----------
TH_1874|M.thermoresistible__bu EVATTVHAHAHLASKLKARDAALKAIRAGIDGIATEFTGG----------
MSMEG_1331|M.smegmatis_MC2_155 EAAA-LHTNAHVASKLKARADAVTAIRAGIDGLAAEFGS-----------
MAB_1069c|M.abscessus_ATCC_199 DDIALGSPQGLAESKRLTTAPILADFDARAVELTETSARLFVSPEAQEGM
MLBr_02118|M.leprae_Br4923 EVTGLAPLAIQHAKRVLNDDGSIEEAWPEHKKLFDKAWTS---QDVIEAQ
: .: : . :
Mb0649c|M.bovis_AF2122/97 --------------
Rv0632c|M.tuberculosis_H37Rv --------------
MMAR_0962|M.marinum_M --------------
MUL_0713|M.ulcerans_Agy99 --------------
MAV_4534|M.avium_104 --------------
Mflv_5125|M.gilvum_PYR-GCK --------------
Mvan_1226|M.vanbaalenii_PYR-1 --------------
TH_1874|M.thermoresistible__bu --------------
MSMEG_1331|M.smegmatis_MC2_155 --------------
MAB_1069c|M.abscessus_ATCC_199 LAFLQKRPPAWQV-
MLBr_02118|M.leprae_Br4923 VARVEKRPPKFQGA