For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTIQDSVAQKQSEVRIQLDRSVLHVLLDRPRKRNALDLTMIRSISRAIDGRPTDTRVVVISGGAFFSAGA DIATYKRGDQGEIGEITRAAGAVIDTMTTAPIPVIAAVEGMALGGGFELAMGADIVVAGESAKLGLPEVA LGLIPGWGGTQRLSAQIGIRRAKQIIMLQQTISAEDAWTLGLVNEVVPDGTSLNRALEMAHQLAASSATA LAATKRLVSGIERNLAYSDERAALMELFGSPDGIEGVTAFVEKRPANFGR
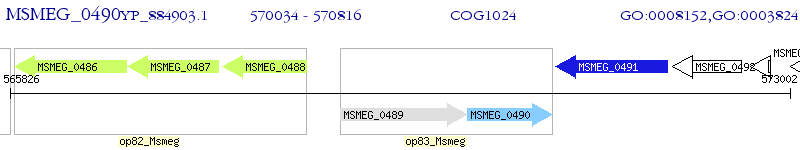
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0490 | - | - | 100% (260) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_6777 | - | 5e-34 | 37.50% (240) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 3e-32 | 36.33% (245) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 1e-32 | 34.36% (259) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 1e-32 | 35.29% (255) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0222 | echA1 | 6e-34 | 32.69% (260) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 7e-29 | 34.96% (246) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_4588c | - | 6e-35 | 37.10% (248) | enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_0465 | echA1 | 4e-33 | 32.27% (251) | enoyl-CoA hydratase EchA1 |
| M. avium 104 | MAV_4951 | - | 5e-34 | 33.47% (251) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_4462 | - | 5e-34 | 35.00% (260) | PUTATIVE putative enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_1114 | echA1 | 8e-33 | 32.27% (251) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4588 | - | 3e-32 | 35.23% (264) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0465|M.marinum_M MSELN----GDTEPEVLVEQRDRILIITINRPKAKNSVNAAVSRALADAM
MUL_1114|M.ulcerans_Agy99 MSELN----GDTEPEVLVEQRDRILIITINRPKAKNSVNAAVSRALADAM
Rv0222|M.tuberculosis_H37Rv MSSESDA--ANTEPEVLVEQRDRILIITINRPKAKNAVNAAVSRGLADAM
MAV_4951|M.avium_104 MRHVSTGNEEAGEPEVLVEQRDRILIITINRPKAKNAVNAAVANGLAAAV
MAB_4588c|M.abscessus_ATCC_199 -----------MADEVLIEQRDRVLLITINRPDARNAVNRAVSQGLAAAA
TH_4462|M.thermoresistible__bu MTAPTTAR--QATDPVLVSADSHVLTITLNRPDVHNAVNLDMAQRISDAL
Mvan_4588|M.vanbaalenii_PYR-1 MTDTVT----DTAPGALTERRGNVLIITINRPEARNAVNASVSIGVGNAL
Mb1099c|M.bovis_AF2122/97 ----------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAA
MLBr_02402|M.leprae_Br4923 ----------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAA
Mflv_2036|M.gilvum_PYR-GCK ----------MTFETILVDRDDRVATITLNRPKALNALNTQVMNEVTTAA
MSMEG_0490|M.smegmatis_MC2_155 --MTIQDSVAQKQSEVRIQLDRSVLHVLLDRPRKRNALDLTMIRSISRAI
. : : ::** *::: : : *
MMAR_0465|M.marinum_M DRLDADAGLSVGILTGAG-GSFCAGMDLKAFARGE---NVVVEGRGLG-F
MUL_1114|M.ulcerans_Agy99 DRLDADAGLSVGILTGAG-GSFCAGMDLKAFARGE---NVVVEGRGLG-F
Rv0222|M.tuberculosis_H37Rv DQLDGDAGLSVAILTGGG-GSFCAGMDLKAFARGE---NVVVEGRGLG-F
MAV_4951|M.avium_104 DRLDSEPGLSVGVLTGAG-GSFCAGMDLKAFARGE---LPIVEGRGMG-F
MAB_4588c|M.abscessus_ATCC_199 DQLDSSADLSVAIITGAG-GNFCAGMDLKAFVSGE---AVLSE-RGLG-F
TH_4462|M.thermoresistible__bu DRLDDDPDLRVGIIAANG-RSFCAGMDLKAFARGE---VPRPEPRGFAGI
Mvan_4588|M.vanbaalenii_PYR-1 QAAQDDPEIRAVILTGAGDKSFCAGADLKAISRGENLFHPEHTEWGFAGY
Mb1099c|M.bovis_AF2122/97 TELDDDPDIGAIIITGSA-KAFAAGADIKEMADLT--FADAFTADFFATW
MLBr_02402|M.leprae_Br4923 KELDIDPDVGAILITGSP-KVFAAGADIKEMASLT--FTDAFDADFFSAW
Mflv_2036|M.gilvum_PYR-GCK AELDAEPGVGAIIVTGSE-RAFAAGADIKEMASLS--FADVFSSDFFAAW
MSMEG_0490|M.smegmatis_MC2_155 DGRPTDT--RVVVISGGA--FFSAGADIATYKRGDQGEIGEITRAAGAVI
.. . :::. *.** *: .
MMAR_0465|M.marinum_M TERPPAK-PLIAAVEGYALAGGTELALATDLIVAARDSAFGIPEVKRGLV
MUL_1114|M.ulcerans_Agy99 TERPPAK-PLIAAVEGYALAGGTELALATDLIVAARDSAFGIPEVKRGLA
Rv0222|M.tuberculosis_H37Rv TERPPTK-PLIAAVEGYALAGGTELALAADLIVAARDSAFGIPEVKRGLV
MAV_4951|M.avium_104 TERPPAK-PLIAAVEGYALAGGTELALATDLIVASKDAAFGIPEVKRGLV
MAB_4588c|M.abscessus_ATCC_199 TNVPPRK-PIIAAVEGFALAGGTELVLSCDLVVAGRSAKFGIPEVKRGLV
TH_4462|M.thermoresistible__bu VKQPPAK-PLVAAVEGAAVGGGFEIVLACDLIVASESARFSLPEVTRGLT
Mvan_4588|M.vanbaalenii_PYR-1 VRHFIDK-PTIAAVNGTALGGGTELALASDLVVAEERAKFGLPEVKRGLI
Mb1099c|M.bovis_AF2122/97 GKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVL
MLBr_02402|M.leprae_Br4923 GKLAAVRTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVL
Mflv_2036|M.gilvum_PYR-GCK GKFAATRTPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVL
MSMEG_0490|M.smegmatis_MC2_155 DTMTTAPIPVIAAVEGMALGGGFELAMGADIVVAGESAKLGLPEVALGLI
* :*** * *:.** *:.: *:::* : :. **: *:
MMAR_0465|M.marinum_M AGGGGLLRLPERIPYAIAMELALTGDNLSAERAHALGMVNVLAEPGAALD
MUL_1114|M.ulcerans_Agy99 AGGGGLLRLPERIPYAIAMELALTGDNLSAERAHALGMVNVLAEPGAALD
Rv0222|M.tuberculosis_H37Rv AGGGGLLRLPERIPYAIAMELALTGDNLPAERAHELGLVNVLAEPGTALD
MAV_4951|M.avium_104 AGGGGLLRLPQRIPSAIAMELALTGDNLSAERAHALGMVNVLAEPGSALD
MAB_4588c|M.abscessus_ATCC_199 AGAGGLLRLPNRIPYQVAMELALTGESFTAEDAAKYGFINRLVDDGQALD
TH_4462|M.thermoresistible__bu PNGGGLLRLPRRIPHHLAMELILTGRPVTAARAAELGLLNRLTPPGSALS
Mvan_4588|M.vanbaalenii_PYR-1 AGAGGVFRIVEQLPRKVALELIYTGEPISSADALRWGLINQVVPDGTVVD
Mb1099c|M.bovis_AF2122/97 PGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLT
MLBr_02402|M.leprae_Br4923 PGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLP
Mflv_2036|M.gilvum_PYR-GCK PGMGGSQRLTRAIGKAKAMDLILTGRNMDAEEAERAGLVSRVVPADSLLS
MSMEG_0490|M.smegmatis_MC2_155 PGWGGTQRLSAQIGIRRAKQIIMLQQTISAEDAWTLGLVNEVVPDGTSLN
.. ** *: : * :: . : * *::. :. . :
MMAR_0465|M.marinum_M AAIALAEKITANGPLAVAATKRIITESRG---WSLDTRFAQQMKILFPIF
MUL_1114|M.ulcerans_Agy99 AAIALAEKITANGPLAVAATKRIITESRG---WSLDTRFAQQMKILFPIF
Rv0222|M.tuberculosis_H37Rv AAIALAEKITANGPLAVVATKRIITESRG---WSPDTMFAEQMKILVPVF
MAV_4951|M.avium_104 EAIALAEKIAANGPLAVAATKRIIVESRG---WSPETMFAEQNKLLAPVF
MAB_4588c|M.abscessus_ATCC_199 TALELAAKITANGPLAVAATKRIIIESAS---WAPEEAFAKQGEILMPIF
TH_4462|M.thermoresistible__bu TARELAAVIAANGPLAIRAAKRVVEESPD---WPRTEMFTRQEDIVAPVR
Mvan_4588|M.vanbaalenii_PYR-1 AALALAERITCNAPLAVQASKRVSYGAVDGVVGSEEPFWKQTFGEFSTLL
Mb1099c|M.bovis_AF2122/97 EARATATTISQMSASAARMAKEAVNRAFES---SLSEGLLYERRLFHSAF
MLBr_02402|M.leprae_Br4923 EAKAVATTISQMSRSATRMAKEAVNRSFES---TLAEGLLHERRLFHSTF
Mflv_2036|M.gilvum_PYR-GCK EAKAVAQTIAGMSLSASRMAKEAVDRAFET---TLSEGLLYERRLFHSAF
MSMEG_0490|M.smegmatis_MC2_155 RALEMAHQLAASSATALAATKRLVSGIER------NLAYSDERAALMELF
* * :: . * :*. .
MMAR_0465|M.marinum_M TSNDAKEGAIAFAEKRPPRWTGT
MUL_1114|M.ulcerans_Agy99 TSNDAKEGAIAFAEKRPPRWTGT
Rv0222|M.tuberculosis_H37Rv TSNDAKEGAIAFAERRRPRWTGT
MAV_4951|M.avium_104 SSNDAKEGAIAFAEKRPPKWTGT
MAB_4588c|M.abscessus_ATCC_199 VSEDAKEGAKAFAEKRAPVWQGK
TH_4462|M.thermoresistible__bu SSADAAEGARAFAEKRMPLWRNR
Mvan_4588|M.vanbaalenii_PYR-1 KTEDAMEGPLAFAQKREPVWKAK
Mb1099c|M.bovis_AF2122/97 ATEDQSEGMAAFIEKRAPQFTHR
MLBr_02402|M.leprae_Br4923 VTDDQSEGMAAFIEKRAPQFTHR
Mflv_2036|M.gilvum_PYR-GCK ATDDQTEGMNAFSEKRSPNFTHR
MSMEG_0490|M.smegmatis_MC2_155 GSPDGIEGVTAFVEKRPANFGR-
: * ** ** ::* . :