For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPPSLSAGPGLRWVLITLCVTEITSWGVLYYAFTVLSASISSDTGWSAPAVTGAFSAGLVTSALMGIVV GRWLDRYGPRWIMTIGSVLGVVAVLAVVVAPNYACFVAAWILAGVAMSGVFYAPAFAALTRYFAPNAVRA LTVLTLVAGLSSTVFAPLTAALGSHLSWRQTYLVLAALLAVITIPAHLFGLRRPWPPVQHHHNVEAPTRT ARSAPFLALMVSFALAALASYAVIVNLVPLMVQRGFSTSAAAVALGLGGAGQVLGRLGYQTMARRIGVVP RTVLIMAAVAVTTALLGVFTTLAALVAASIVAGMARGIMTLLQATAVTDRWGTTHYGHLSGMLMAPIMIT TALGPFVGAALASLLDGYDAMFLVLGVVAAVAAALAAATGVRPTSGSADPSAAGDRRPPCSAEH
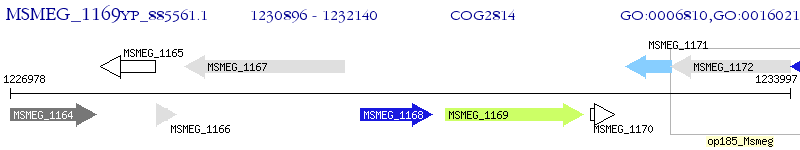
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1169 | - | - | 100% (414) | integral membrane transport protein |
| M. smegmatis MC2 155 | MSMEG_5464 | - | 1e-68 | 40.53% (375) | integral membrane transport protein |
| M. smegmatis MC2 155 | MSMEG_4104 | - | 6e-33 | 28.24% (393) | transporter, major facilitator family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 2e-09 | 22.70% (326) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1096 | - | 7e-22 | 26.78% (407) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0191 | - | 2e-09 | 22.70% (326) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4839c | - | 1e-149 | 67.55% (379) | MFS family transporter |
| M. marinum M | MMAR_0577 | - | 2e-14 | 28.99% (414) | integral membrane transport protein |
| M. avium 104 | MAV_4987 | - | 3e-13 | 25.00% (360) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_0476 | - | 3e-13 | 25.07% (363) | major facilitator superfamily protein MFS_1 |
| M. ulcerans Agy99 | MUL_1084 | - | 4e-11 | 22.47% (356) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_3219 | - | 1e-161 | 73.23% (381) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1169|M.smegmatis_MC2_155 MTPPSLSAGPG----LRWVLITLCVTEITSWGVLYYAFTVLSAS--ISSD
Mvan_3219|M.vanbaalenii_PYR-1 MAE---SDRAD----RRSVLLTLCLTEITSWGVLYYAFSVLSPA--ICAD
MAB_4839c|M.abscessus_ATCC_199 MSTAAVHAQPHSITGLRGVLITLCLTEITSWGVLYYAFPVLAPR--ITAD
Mb0197|M.bovis_AF2122/97 MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGALSAIARN
Rv0191|M.tuberculosis_H37Rv MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGALSAIARN
MUL_1084|M.ulcerans_Agy99 MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARN
MAV_4987|M.avium_104 MTAET--ANAAARTWTPRIAAQLAILAAAAFTYVTAEILPVGALPAIARN
MMAR_0577|M.marinum_M -MPESVTAPPELG--IRHATPGLLALTIATCLAITTEILPVGLLPAIGHT
Mflv_1096|M.gilvum_PYR-GCK MGGPVTRFDSAAIAEWRRYGTVPLAAGLGYSMGVIHVYSLGAFMGPLQDE
TH_0476|M.thermoresistible__bu ---------------VRRFAFPLLAYAFAAVMVGTTVPTPMYAV--YAER
MSMEG_1169|M.smegmatis_MC2_155 TGWSAPAVTGAFSAGLVTSALMGIVVGRWLDRYGPRWIMTIGSVLGVVAV
Mvan_3219|M.vanbaalenii_PYR-1 TGWSAPAVTAAFSAGLVISALLGIPVGRWLDRHGPRWIMTGGSVLGSIAL
MAB_4839c|M.abscessus_ATCC_199 TGWSAPAVTAAFSAALVVAAAAGIGVGRWLDRHGPRWLMTAGSLWGPAAL
Mb0197|M.bovis_AF2122/97 LRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQ
Rv0191|M.tuberculosis_H37Rv LRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQ
MUL_1084|M.ulcerans_Agy99 LHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHALMVSLVCLTISQ
MAV_4987|M.avium_104 LQVSLVLVGTLLSWYALVAALTTIPLVRWTAHLPRRRVLVASLTCLTASQ
MMAR_0577|M.marinum_M FGVRDAVTGLLVSLYAVLVALLAVPLTVVTARFPRKPLLLTTLLGYAVSN
Mflv_1096|M.gilvum_PYR-GCK FGWSRAQASVGLTIVGMAAAVAALPIGLMVDRLGPRRVGIIGAGLMAGAF
TH_0476|M.thermoresistible__bu LGFGVLTTTVVFAGYTAGVLFALVVFGRWSDALGRRPMLLAGVGCAIVSA
. .: : . : :
MSMEG_1169|M.smegmatis_MC2_155 LAVVVAP-NYACFVAAWILAGVAMSGVFYAPAFAALTRYFAPNAVRALTV
Mvan_3219|M.vanbaalenii_PYR-1 AGVAAAP-DVVSFTAAWVVAGVAMSAVFYAPAFAALTRYFRPDAVRALTA
MAB_4839c|M.abscessus_ATCC_199 IGVAMAP-NYGWFLAAWLMAGIAMAAVFYPPAFAALTRWYGPGAVRALMV
Mb0197|M.bovis_AF2122/97 LVSALAP-NFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTS
Rv0191|M.tuberculosis_H37Rv LVSALAP-NFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTS
MUL_1084|M.ulcerans_Agy99 LISALAP-NFAVLAAGRALCAITHGLLWSVIAPIATRLVPPSHAGRATTS
MAV_4987|M.avium_104 LISALAP-NFAVLAAGRVLCAITHGLLWSVIAPIATRLVPPSHAGRATMS
MMAR_0577|M.marinum_M TLVAIAP-TFAMVAAGRAVGGVTHALFFSLCIGYAPRLVGLADVGRALAL
Mflv_1096|M.gilvum_PYR-GCK ALLGTATGTMGNWILLWSVLAFACFWVQTTVWTSAVASRFETSRGLAFAV
TH_0476|M.thermoresistible__bu LVFLIAG-SVPQLIAARFVSGMSAGIFTGTATAAVIEAAPERRRSRAAAV
* : ..: . . *
MSMEG_1169|M.smegmatis_MC2_155 LTLVAGLSSTVFAPLTAALGSHLSWRQTYLVLAALLAVITIPAHLFGLRR
Mvan_3219|M.vanbaalenii_PYR-1 LTLAAGLASTVFAPLTAAISAHMDWRETYLVLAAAMAVITVPAHFFGLRR
MAB_4839c|M.abscessus_ATCC_199 LTLVAGLASTVFVPLTAVLASHMDWRTTYLALAVLLTIITVPAHIIGLRR
Mb0197|M.bovis_AF2122/97 IYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAARLA-LPE
Rv0191|M.tuberculosis_H37Rv IYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAARLA-LPE
MUL_1084|M.ulcerans_Agy99 IYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVVTVAARLL-LPE
MAV_4987|M.avium_104 IYVGTSLALVVGSPLTAALSLMWGWRLAVVCVTVAAAVVTVAARLM-LPE
MMAR_0577|M.marinum_M VGGGVSAGIVVGVPLLTSVGTAVGWRGAFAVLAVLAAVL-----LLVVAK
Mflv_1096|M.gilvum_PYR-GCK TLSGGSLAAAVFPPLATLLIGEWGWRGALFGLGGLWGGLVLIVVLLFFRG
TH_0476|M.thermoresistible__bu ATVANIGGLGAGPLLAGILVQYAPWPLHLTFWVHIVLMVLAAAAIAVAPE
. * : * : :
MSMEG_1169|M.smegmatis_MC2_155 PWP---PVQHHHNVEAPTRTA-----------RSAPFLALMVSFALAALA
Mvan_3219|M.vanbaalenii_PYR-1 PWP---HIEHHHLVEPPTRIA-----------RSRQFVALTVAFAMAGLA
MAB_4839c|M.abscessus_ATCC_199 PWP---SLPATHLAESPTRIA-----------RSRPFISLIIAFALASCA
Mb0197|M.bovis_AF2122/97 MVL---RADQLEHVGRRARHH-----------RNPRLVKVSVLTMIAVTG
Rv0191|M.tuberculosis_H37Rv MVL---RADQLEHVGRRARHH-----------RNPRLVKVSVLTMIAVTG
MUL_1084|M.ulcerans_Agy99 MVL---SADQLQHVGPRSRHH-----------RNRALIIVSLITMVGVTG
MAV_4987|M.avium_104 MVL---TEHQLAHVGPRSRHH-----------RNGRLITVSLLAMVAVTG
MMAR_0577|M.marinum_M LLP---PVSSESAPPPAGTAK-----------GRRRLAAVVSSNALTYTG
Mflv_1096|M.gilvum_PYR-GCK AQDDKKPAPARTVIAPTAAAADLPGVTLAEGLRSATFYKLLVAAGLFAFT
TH_0476|M.thermoresistible__bu TSP----RTGRLGVQRLSLPP-----------QVRKVFAIASLAAFAGFA
. : .
MSMEG_1169|M.smegmatis_MC2_155 SYAVIVNLVPLMVQRGFSTSAAAVALGLGGAGQVLGRLGYQTMARRIG--
Mvan_3219|M.vanbaalenii_PYR-1 SYAVIVNLVPLMTAHGISTQAAALALGLGGAGQVLGRLGYQRMVRHLS--
MAB_4839c|M.abscessus_ATCC_199 SYAVIVNLVPLMNQRGLDGTVAATALALGGVGQVIGRFGYPALAQCIS--
Mb0197|M.bovis_AF2122/97 HFVSYTYIVVIIR-DVVGVRGPNLAWLLAAYG-VAGLVSVPLVARPLD--
Rv0191|M.tuberculosis_H37Rv HFVSYTYIVVIIR-DVVGVRGPNLAWLLAAYG-VAGLVSVPLVARPLD--
MUL_1084|M.ulcerans_Agy99 HFVSYTYIVVIIR-QVVGVRGPSLAWLLAAYG-VAGVVAVALVARPLD--
MAV_4987|M.avium_104 HFVSYTFIVEIIR-NVVGVRGPTLAWVLAAYG-LAGLLSVPLVARPLD--
MMAR_0577|M.marinum_M QFTLYTFISAVLL--AAGARPAFIGPLLLMCG-VCSLAGLWYVGATLD--
Mflv_1096|M.gilvum_PYR-GCK TLGIVVHLVPILRDAGAEPLAAAGTAALVGIFSIVGRLGVGLLLDRFPG-
TH_0476|M.thermoresistible__bu VTGLFTAVAPSFLAEVIGVRNHAVAGAVAASIFAASAVTQVLGASVSPRR
. : : : .
MSMEG_1169|M.smegmatis_MC2_155 VVPRTVLIMAAVAVTTALLGVFTTLAALVAASIVAGMARGIMTLLQATAV
Mvan_3219|M.vanbaalenii_PYR-1 VVPRTALIMAGVAVTTALLGLLTSVATLITVAVVAGVVRGVMTLLQATAV
MAB_4839c|M.abscessus_ATCC_199 VVPRTLLIMGVVAATTALLGVFASAVSLIAVAVAAGMARGIMTLLQATAI
Mb0197|M.bovis_AF2122/97 RWPKGAVIVGMTGLTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALA
Rv0191|M.tuberculosis_H37Rv RWPKGAVIVGMTGLTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALA
MUL_1084|M.ulcerans_Agy99 RRPKGTIIFCVAGLTFAFVLLTALAFGGHLAPMTALVVGTGAIVLWGAAV
MAV_4987|M.avium_104 HRPKSAVILCMTGLTAAFAVLTALAFGGPTGATTALIG-TAAIVLWGAMA
MMAR_0577|M.marinum_M RNPRQTTVIIIVVVIGALLGLG--------ASFPALLPVVLAAAVWNGAF
Mflv_1096|M.gilvum_PYR-GCK RVVGACAYLVPILGCAALLIDGSSPLSQTVAAALFGLTLGAEVDVIAYLA
TH_0476|M.thermoresistible__bu AVAVGCALLIVGMVFLAAALVWSSLPALIATAVVAGAGQGLSFSRGLAAV
. * .
MSMEG_1169|M.smegmatis_MC2_155 TDRWGTTHYGHLSGMLMAP------IMITTALGPFVGAALAS-LLDGYDA
Mvan_3219|M.vanbaalenii_PYR-1 TERWGITHYGQLSGILSAP------VMLSTALGPFVGAGLAT-LLGGYGA
MAB_4839c|M.abscessus_ATCC_199 TERWGSTHYGHLSGLLTAP------IMLTTALAPFTGAALAQ-LLHGYPA
Mb0197|M.bovis_AF2122/97 TAVSPMLQSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGG-LLYERSL
Rv0191|M.tuberculosis_H37Rv TAVSPMLQSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGG-LLYERSL
MUL_1084|M.ulcerans_Agy99 TAVSPMLQSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGG-LLYERSV
MAV_4987|M.avium_104 TAVSPMMQSAAMRNGADDPDGASGLYVTAFQVGIMAGSLLGG-LLYERSV
MMAR_0577|M.marinum_M GGIPSIYQAGAVRAQAATPEMAGAWVNATANLGIAAGAAIGAGLLPTIGL
Mflv_1096|M.gilvum_PYR-GCK SRYFGLRNFGGLFGGLVAA------LSLGTAFGPLAAGATSDHFGGYTNF
TH_0476|M.thermoresistible__bu SARTPSDQRGEVSSTYFVV-------AYVALLVPVIGEGWAAHHWGLRTA
: . : . . .
MSMEG_1169|M.smegmatis_MC2_155 MFLVLGVVAAVAAALAAATGVRPTSGSADPSAAGDRRPPCSAEH
Mvan_3219|M.vanbaalenii_PYR-1 MFVVLGAVALAAALLATATRVRPVDADG----------------
MAB_4839c|M.abscessus_ATCC_199 MFVALGVAGIIGAAAATAT--NPSQ-------------------
Mb0197|M.bovis_AF2122/97 AMMLTASAGLMGVALFGMTVSQHLFENPTLSPGDG---------
Rv0191|M.tuberculosis_H37Rv AMMLTASAGLMGVALFGMTVSQHLFENPTLSPGDG---------
MUL_1084|M.ulcerans_Agy99 AMMLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ-----
MAV_4987|M.avium_104 ILMLSASGVLMAVALVGIAANRRMLDVAPTSSRDS---------
MMAR_0577|M.marinum_M AGLPWVAAALIAISLAVTLATGRGPTH-----------------
Mflv_1096|M.gilvum_PYR-GCK LILTMVLMGLSSMALASLKPPPVWTVTSPGEDARADRFPQP---
TH_0476|M.thermoresistible__bu GISFAVGVAVLAIGCLIAILIQESRDPQAI--------------
.