For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPGPDAASRAILDAALVEFERYGFRRVALDDVARRAGVSRTTIYRRFANRDDLVGAVIERENVSLFADIA AELKDAKPQSNIYVEAFTLSILRFRRHRVLNQMMKDEPALVLELAHRHYGAAIERMAAALRVIFPEGFAD RIGEAAVNDLADTILRYALMALLIPSVEPLETADDIRSFATRHFLPSLPVSLRVVPA
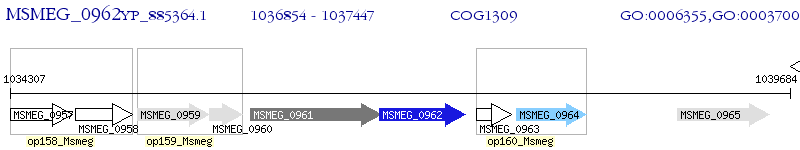
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0962 | - | - | 100% (197) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1500 | - | 1e-08 | 46.55% (58) | TetR family protein transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_5040 | - | 5e-08 | 24.18% (182) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0237 | - | 2e-07 | 28.00% (125) | TetR/ACRR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0059 | - | 5e-81 | 77.16% (197) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0232 | - | 2e-07 | 28.00% (125) | TetR/ACRR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00815 | - | 3e-05 | 32.14% (56) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1024 | - | 1e-19 | 30.26% (195) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0287 | - | 3e-21 | 34.38% (192) | transcriptional regulatory protein |
| M. avium 104 | MAV_5227 | - | 3e-20 | 35.00% (180) | TetR-family protein transcriptional regulator, putative |
| M. thermoresistible (build 8) | TH_2255 | - | 1e-78 | 74.60% (189) | TetR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_4837 | - | 1e-21 | 34.90% (192) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0855 | - | 9e-82 | 78.17% (197) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0059|M.gilvum_PYR-GCK --------------------------------MS-GPDPASEQILDAAVV
Mvan_0855|M.vanbaalenii_PYR-1 --------------------------------MR-GTD-ATEHILDAAVV
MSMEG_0962|M.smegmatis_MC2_155 --------------------------------MP-GPDAASRAILDAALV
TH_2255|M.thermoresistible__bu --------------------------------MPERPDPARQAILDAAVV
MMAR_0287|M.marinum_M --------------------MQSRMGTETSTDGVESITDLDRTILDTARA
MUL_4837|M.ulcerans_Agy99 ------------------------MGTETSTDGVESITDLDRTILDTAHA
MAV_5227|M.avium_104 ------------------------MQSHRADAAAGERTDLDRLILDTARG
Mb0237|M.bovis_AF2122/97 ---------------------------MPTVTWARVDPARRAAVVEAAEA
Rv0232|M.tuberculosis_H37Rv ---------------------------MPTVTWARVDPARRAAVVEAAEA
MAB_1024|M.abscessus_ATCC_1997 --------------------------------MSADPDPTGERILEAALQ
MLBr_00815|M.leprae_Br4923 MNNLAKAAQRRTVRPIDRGRLSDDIATPNQRSNRLSRDKRRGQLLIVASD
:: .*
Mflv_0059|M.gilvum_PYR-GCK EFERHGFRRVALEDVARRAGVSRTTIYRRFANKD---QLVGAVIERENVA
Mvan_0855|M.vanbaalenii_PYR-1 EFERHGFRRVALDDVARRAGVSRTTIYRRFANKD---ELVGAVIERENIQ
MSMEG_0962|M.smegmatis_MC2_155 EFERYGFRRVALDDVARRAGVSRTTIYRRFANRD---DLVGAVIERENVS
TH_2255|M.thermoresistible__bu EFEQHGFRKVALEDVARRAGVSRTTIYRRFTNRD---ELVAAVVDRENAA
MMAR_0287|M.marinum_M VFETYGVRRANIEDVATRAGVSRSTIYRRFPTKD---ELFERVVRREAEL
MUL_4837|M.ulcerans_Agy99 VFETYGVRRANIEDVATRAGVSRSTIYRRFPTKD---ELFERVVRREAEL
MAV_5227|M.avium_104 VFETYGVRRANIDDVAARAGVSRSTIYRRFPTKK---NLFGQVVRREAEL
Mb0237|M.bovis_AF2122/97 EFGAHGFSRGSLNVIARRAGVAKGSLFQYFADKRDLYAFIADIASQRVRS
Rv0232|M.tuberculosis_H37Rv EFGAHGFSRGSLNVIARRAGVAKGSLFQYFADKRDLYAFIADIASQRVRS
MAB_1024|M.abscessus_ATCC_1997 TMLSFGIRRATVDEIARRAGVSHMTVYRRWSNKT---DLVLAVLMREAQA
MLBr_00815|M.leprae_Br4923 VFVDHGYHAAGMDEIADRAGVSKPVLYQHFSSKL---ELYLAVLERHMNN
: .* :: :* ****:: ::: :. : : : :.
Mflv_0059|M.gilvum_PYR-GCK LFSDIATELKQAGPQSNYYVEAFTLSILKFRGHRVLHRMITEEPGLVLEL
Mvan_0855|M.vanbaalenii_PYR-1 LFADIAAELKTAGPQSNYYVEAFTLSILKFRRHRVLHQMIIDEPGLVLEL
MSMEG_0962|M.smegmatis_MC2_155 LFADIAAELKDAKPQSNIYVEAFTLSILRFRRHRVLNQMMKDEPALVLEL
TH_2255|M.thermoresistible__bu LFADIAAELKHSGPQGDYYVEAFTLAILKFRRHRVLHRMITDEPGFILHL
MMAR_0287|M.marinum_M FFAALNQATNGHNPR-EAVIEAFTLGVRLIQDSPLYSRIAESEP-ELFGM
MUL_4837|M.ulcerans_Agy99 FFAALNQATNGHNPR-EAVIEAFTLGVRLIRDSPLYSRIAESEP-ELFGM
MAV_5227|M.avium_104 FFSTLDQATAGCDPQ-QAVIEAFALGVRLVKNSALYSRIAESEP-ELFGL
Mb0237|M.bovis_AF2122/97 YMEDLIRELDPNRPFFEFLTDLLDGWVAYFAEHPRERALHAAATLEVDTD
Rv0232|M.tuberculosis_H37Rv YMEDLIRELDPNRPFFEFLTDLLDGWVAYFAEHPRERALHAAATLEVDTD
MAB_1024|M.abscessus_ATCC_1997 IFTSVDAEIAALDSPEEKLVAGFTGIYWHVHTHPLMRRAVETDPESVLPV
MLBr_00815|M.leprae_Br4923 LVSSVQQALRTTTNNRQRLHAAVQAFFDFIEHDGQGYRLIFENDNITEPR
. : : . .
Mflv_0059|M.gilvum_PYR-GCK ASLYYRAAIER-MAEALRVIFPPGFA--ERIGADAVDELADTILRYATMV
Mvan_0855|M.vanbaalenii_PYR-1 AGRYHRAAIDR-MAAALRVIFPPGFA--ERIGPAAVDELADCILRYAAMV
MSMEG_0962|M.smegmatis_MC2_155 AHRHYGAAIER-MAAALRVIFPEGFA--DRIGEAAVNDLADTILRYALMA
TH_2255|M.thermoresistible__bu AQQHWGAAVER-MAEALRVIFPAGFA--ERIGEAAVIEFADTILRYAAMA
MMAR_0287|M.marinum_M FSRSHVFPIGQ-FADGIAHTLRRCGA--DAP-DADLANIADILLRVAVGI
MUL_4837|M.ulcerans_Agy99 FSRSHVFPIGQ-FADGIAHTLRRCGA--DAP-DADLANIADILLRVAVGI
MAV_5227|M.avium_104 FSRSQVFPIRQ-FADGIAHTLRRCGA--EQA-EPDLDNIADILLRVALGI
Mb0237|M.bovis_AF2122/97 ARISVRSVLHRHYLDVLRPLVRDAHARGDLRADSDTGALMSLLLLIFPHL
Rv0232|M.tuberculosis_H37Rv ARISVRSVLHRHYLDVLRPLVRDAHARGDLRADSDTGALMSLLLLIFPHL
MAB_1024|M.abscessus_ATCC_1997 MTSGAGPALDMATTYLAGHVSQSAGD-----LVDDPYGIAEVFVRLTHSL
MLBr_00815|M.leprae_Br4923 VAAQVRVATESCIDAVFALISADSRLD---PHRARMVAVGLVGISVDCAR
. :
Mflv_0059|M.gilvum_PYR-GCK LLLPSAQPLETAD-------DIRAFARHHFLPSLPEALRSVPA-------
Mvan_0855|M.vanbaalenii_PYR-1 LLLPSAQPLETAD-------DVRAFARQHFLPSLPAALRTVPA-------
MSMEG_0962|M.smegmatis_MC2_155 LLIPSVEPLETAD-------DIRSFATRHFLPSLPVSLRVVPA-------
TH_2255|M.thermoresistible__bu LLLPSVQPLDTAD-------DIRAFATRHFLPSLPAALHTGSA-------
MMAR_0287|M.marinum_M IMFPTER-LDTSDDA-----AVREYAARYLIPIITL--------------
MUL_4837|M.ulcerans_Agy99 IMFPTER-LDTSDDA-----AVREYAARYLIPIITL--------------
MAV_5227|M.avium_104 IVFPTDR-LDTADPA-----AVRDYAARYLVPIIGR--------------
Mb0237|M.bovis_AF2122/97 ALAPYMRGLDPILGLDEPTPEQPALAVRRLVAVLAAAFDAQHPATNSAQT
Rv0232|M.tuberculosis_H37Rv ALAPYMRGLDPILGLDEPTPEQPALAVRRLVAVLAAAFDAQHPATNSAQT
MAB_1024|M.abscessus_ATCC_1997 ILAPSPRRELATR------EDAQQYARRYILPIAQALVPAASPAVR----
MLBr_00815|M.leprae_Br4923 YWLDTNRPISKSD-------AVEGTVQFAWGGLSHVPLTRL---------
. .
Mflv_0059|M.gilvum_PYR-GCK ------
Mvan_0855|M.vanbaalenii_PYR-1 ------
MSMEG_0962|M.smegmatis_MC2_155 ------
TH_2255|M.thermoresistible__bu ------
MMAR_0287|M.marinum_M ------
MUL_4837|M.ulcerans_Agy99 ------
MAV_5227|M.avium_104 ------
Mb0237|M.bovis_AF2122/97 RSEEIT
Rv0232|M.tuberculosis_H37Rv RSEEIT
MAB_1024|M.abscessus_ATCC_1997 ------
MLBr_00815|M.leprae_Br4923 ------