For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLLNWKYVESRADDDFVVAPGERLDWGRTIGIGAQHVVAMFGATFLVPVLTGFPPSTTLLFSGIGTVLFL LITGNRLPSYLGSSFAVIAPVTAAVAADGAGSALGGLIAVGVLLIIVGAVVHLVGTHWIDVVLPPVVTGA IVALIGFNLAPAAKNNFEQGPLVGLVTLVLLVGVLAFFRGIIGRLAIFGAVVIGYLLALVLGEVDTSAIA AAPWVGLPQFQTPTFSLGLLPLFLPAVIALIAENIGHVKSVGQMTKTDMDPLIGRALAADGVATVLAGAG GGSATTTYAENIGVMAATRVYSTAAYWVAAVVAIVLSLCPKVGAVISAIPAGVLGGATIVLYGLVGILGV RIWLTNHVDFSKPVNQMTAAIPLIIGIADFTWQAGELTFTGIALGSIAALAVFHGMRLLETLGGLGRRPA ADQAPTPPTH*
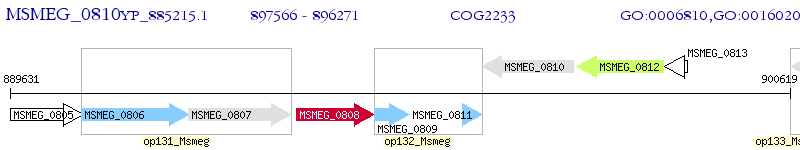
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0810 | - | - | 100% (431) | putative pyrimidine permease RutG |
| M. smegmatis MC2 155 | MSMEG_4011 | - | e-160 | 67.96% (412) | putative pyrimidine permease RutG |
| M. smegmatis MC2 155 | MSMEG_2570 | - | 1e-36 | 28.74% (421) | xanthine/uracil permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0188 | - | 0.0 | 81.60% (424) | uracil-xanthine permease |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2932c | - | 2e-36 | 27.64% (398) | xanthine/uracil permease |
| M. marinum M | MMAR_4949 | - | 4e-35 | 28.33% (413) | xanthine/uracil permease |
| M. avium 104 | MAV_1023 | - | 1e-06 | 23.32% (386) | MFS transporter |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4272 | - | 8e-05 | 26.20% (271) | hypothetical protein MUL_4272 |
| M. vanbaalenii PYR-1 | Mvan_0717 | - | 0.0 | 84.44% (405) | uracil-xanthine permease |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2932c|M.abscessus_ATCC_199 MSADTTRPAAPGAQRKKVHPVDEVPSTGRLVTLGIQHVIAFYAGAVLVPL
MMAR_4949|M.marinum_M MSAPPTSPVR------RVHPVDEVLPIPKLATYGFQHVVAFYAGAVVVPI
Mflv_0188|M.gilvum_PYR-GCK -MTLTWKRVDSTTDPDFVVAPDERLSWPSTLGLGAQHVVAMFGATFLVPV
Mvan_0717|M.vanbaalenii_PYR-1 --MRAWTKVDSRPDPDFVVAPDERLSWPRTLGLGAQHVVAMFGATFLVPV
MSMEG_0810|M.smegmatis_MC2_155 MTLLNWKYVESRADDDFVVAPGERLDWGRTIGIGAQHVVAMFGATFLVPV
MUL_4272|M.ulcerans_Agy99 --MSTLPPPGYPVEPNSGAPGYEAAGYP---PSGYADPPPAYGPPPAYGP
MAV_1023|M.avium_104 -----------------MTATPRACNRDRVALQAVHFFMADMEAG-----
. . . .
MAB_2932c|M.abscessus_ATCC_199 LIARAIDLDAEALTMLITADLFTCGVASILQAVGFWKIGVRLPLLQGVTF
MMAR_4949|M.marinum_M IIANAIDLPHQDLVKLITADLFTCGVASIIQSVGFWKIGVRLPLLQGVAF
Mflv_0188|M.gilvum_PYR-GCK LTG-----------FPPATTLLFSGVGTILFLL---ITGNRLPSYLGSSF
Mvan_0717|M.vanbaalenii_PYR-1 LTG-----------FPPATTLLFSGVGTILFLL---ITGNRLPSYLGSSF
MSMEG_0810|M.smegmatis_MC2_155 LTG-----------FPPSTTLLFSGIGTVLFLL---ITGNRLPSYLGSSF
MUL_4272|M.ulcerans_Agy99 PPGYG---------APPPGYGPPPGYAPPPPGYGP-PPGYGPPPGYGPPP
MAV_1023|M.avium_104 ---------------------MGPFLGVLLQSRGWTTGAIGAAMTLGAIV
. . . *
MAB_2932c|M.abscessus_ATCC_199 ATLAPVIKIANDHGGGRVG--LLTVYGAVIAAGAFTFLIAPFFARLIRFF
MMAR_4949|M.marinum_M AGVSPVIAIGLSHGGGTPS--LLYVYGAVIVAGLFTILIAPIFTKLLRFF
Mflv_0188|M.gilvum_PYR-GCK SVIAPVTAATASHGTGSALGGIIAVGVLLIIIGAVVHVAGTRWIDIT--L
Mvan_0717|M.vanbaalenii_PYR-1 SVIAPVTAAVASEGTGSALGGIVAVGVLLILVGGVVHMVGTRWLDLT--L
MSMEG_0810|M.smegmatis_MC2_155 AVIAPVTAAVAADGAGSALGGLIAVGVLLIIVGAVVHLVGTHWIDVV--L
MUL_4272|M.ulcerans_Agy99 PGYGPPLVPGAVKPG---IIPLRPLTLSDIFNGAVGYIRANPKATLG---
MAV_1023|M.avium_104 GMVTVAPAGALVDATRHKRGCVIVVGLAAVAASAVILTSRQFWTVATAQA
. : : : . .
MAB_2932c|M.abscessus_ATCC_199 PPVVTGSVIT--IIGVSLLPVGIGDATRNPHSPVAAHDPGNGRWLMYALG
MMAR_4949|M.marinum_M PPVVTGTLIT--IIGLCLVPVGAGDAVTNPRT--GEHDPTNIRWLLYALG
Mflv_0188|M.gilvum_PYR-GCK PPVVTGAIVA--LIGFNLAPAAKNNFEQGPLLG---------------FV
Mvan_0717|M.vanbaalenii_PYR-1 PPIVTGAIVA--LIGFNLAPAAKNNFEQGPLLG---------------LV
MSMEG_0810|M.smegmatis_MC2_155 PPVVTGAIVA--LIGFNLAPAAKNNFEQGPLVG---------------LV
MUL_4272|M.ulcerans_Agy99 ---LTAIVVV--VMQVITLLAAIGPLEAASRFD---------------TH
MAV_1023|M.avium_104 VMCISGATIAPAMIGITLGVVGQAAFTRQNGRN------------QAYNH
::. :. :: . ..
MAB_2932c|M.abscessus_ATCC_199 TIGAIVLMQRFFRGFWSTIAVLLGLVAGTVVAWLAGDTDFGRVGEADWIG
MMAR_4949|M.marinum_M TIAIIVAIQRVFSGFMATIAVLIGLVVGCVVAYGLGDMSFAKVDEASVIG
Mflv_0188|M.gilvum_PYR-GCK TLVLLVVVLAAFRGLLGRLAIFVAVAAGYLLAVFLGEVDTSGIAAAAWIG
Mvan_0717|M.vanbaalenii_PYR-1 TLVLLVAALAFFRGMIGRLAIFVAVATGYVLALLLGEVDTSGIATAPWIG
MSMEG_0810|M.smegmatis_MC2_155 TLVLLVGVLAFFRGIIGRLAIFGAVVIGYLLALVLGEVDTSAIAAAPWVG
MUL_4272|M.ulcerans_Agy99 PSELSWGVVGAWTG-SAAAGLLVSWLGGMLLSGILTVVVGRAVFGSPITI
MAV_1023|M.avium_104 AGNMAGAALGGVAGWVFGYAGIFWLAAGFAVATIAAVLAIPGGDIDHHVA
. * . : * ::
MAB_2932c|M.abscessus_ATCC_199 FTSPFAFGAPRFDVIAVVSLIVVLMVTAVESTGAVFATAEIVGKRVRGAD
MMAR_4949|M.marinum_M FTEPFVFGMPKFDFVACLTMIIVLMITSVESTGSTLATGEIVGKRIKAAQ
Mflv_0188|M.gilvum_PYR-GCK LPD---FQAPTFTLSVLPLFLPAVVALVAENIGHVKSVGQMT-KRDMDPL
Mvan_0717|M.vanbaalenii_PYR-1 LPE---FEAPTFTLSVLPMFLPAVIALIAENIGHVKSVSQMT-KTDLDPL
MSMEG_0810|M.smegmatis_MC2_155 LPQ---FQTPTFSLGLLPLFLPAVIALIAENIGHVKSVGQMT-KTDMDPL
MUL_4272|M.ulcerans_Agy99 SEAWARVRGRILALIGLALLEGAGLVALAGLVVVILVGVAAAANGATAAL
MAV_1023|M.avium_104 RGEARAAGEAPVKAMRVLARSRPLLVLAAAMVLFHLGNAAMLPLYGLAVV
. : .
MAB_2932c|M.abscessus_ATCC_199 IAATVRADGIATVIGGTFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVI
MMAR_4949|M.marinum_M IGNVLRADGLATVIGGIFNSFPYTAFSENVGLVRLTRVKSRWVVAAAGVV
Mflv_0188|M.gilvum_PYR-GCK TGRALAADGVATTLAGFGGGSATTTYAENIGVMAATRVYSTAPYWIAALV
Mvan_0717|M.vanbaalenii_PYR-1 MGRALAADGVATTLAGFGGGSATTTYAENIGVMAATRVYSTAAYWVAALV
MSMEG_0810|M.smegmatis_MC2_155 IGRALAADGVATVLAGAGGGSATTTYAENIGVMAATRVYSTAAYWVAAVV
MUL_4272|M.ulcerans_Agy99 LGFPLVLL-VGLLVAYLYTVLMFAPVLIVLERLPLVDAITRSFALVTGGF
MAV_1023|M.avium_104 ATHANPFTTVASTVVVAQAVMVPASLLAMR--IAATRGYWPAILIALTAL
:. : :. : . .
MAB_2932c|M.abscessus_ATCC_199 MMVLGVLPKTAKIVESIPAPVLG--GAAMTLFATVAVVGIQTLGKVDFND
MMAR_4949|M.marinum_M MIVLGFLPKVAAVVSAVPSPVLG--GAALTLFATVAVVGIQTLGKVDFTD
Mflv_0188|M.gilvum_PYR-GCK AILLSLCPKVGAVIAAIPAGVLG--GATVVLYGLVGIIGIR----IWLTN
Mvan_0717|M.vanbaalenii_PYR-1 AVALSLCPKVGAVIVAVPAGVLG--GATVVLYGLVGILGVR----IWLTN
MSMEG_0810|M.smegmatis_MC2_155 AIVLSLCPKVGAVISAIPAGVLG--GATIVLYGLVGILGVR----IWLTN
MUL_4272|M.ulcerans_Agy99 WRVLGIR-----LLTAIVVGLVG--GAISAPFGIVGQILLG----ATASE
MAV_1023|M.avium_104 PVRGVLAASVITSWGVIPVQVLDGIGAGMLSVAVPGLVARILDGTGHINV
. : ::. ** . . : .
MAB_2932c|M.abscessus_ATCC_199 HRNLIIASTSLAMGMYVQFSQSSGPATVLDHGRGIAVPALPGIDQAVPGL
MMAR_4949|M.marinum_M HRNLIIVTTSLALALWVTS--------------------YPDVAESMPSG
Mflv_0188|M.gilvum_PYR-GCK HVDFSKPVN-----------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 HVDFSKPIN-----------------------------------------
MSMEG_0810|M.smegmatis_MC2_155 HVDFSKPVN-----------------------------------------
MUL_4272|M.ulcerans_Agy99 GSTGMFLVG-----------------------------------------
MAV_1023|M.avium_104 GQGAVMAAQG----------------------------------------
MAB_2932c|M.abscessus_ATCC_199 LQIPFSTGITMGAITAIVLNLLFFHTGSRGVAVAGGGAIRLAQVNEMTLE
MMAR_4949|M.marinum_M VDLIFGSGISIGAVAAIVLNIVFFHTGSHGPRVAGGGSITLHEVNAMTRE
Mflv_0188|M.gilvum_PYR-GCK ---------QMTAAIPLIIGIADFTWSAGSLTFTG---IALGSIAALLIY
Mvan_0717|M.vanbaalenii_PYR-1 ---------QMTAAIPLIIGIADFTWSAGGLTFTG---IALGSIAALLIY
MSMEG_0810|M.smegmatis_MC2_155 ---------QMTAAIPLIIGIADFTWQAGELTFTG---IALGSIAALAVF
MUL_4272|M.ulcerans_Agy99 --------MTLSSIGSAISQIITAPFTAGVVVLLY----TDRRMRAEAFD
MAV_1023|M.avium_104 ---------LGGALSPVLGGAVAQHLGFRAAFLLLAG-LSLGALIIWVTF
: . : . :
MAB_2932c|M.abscessus_ATCC_199 QFRRTFGGLVQDVHWVVDRAYRQRPFADAHDLRSAFQEAMLTGSDEEQLE
MMAR_4949|M.marinum_M RFTEVFGHVVQDVPWVIDRAYEHRPFADTAALREAFQDAVLTGSSEQQME
Mflv_0188|M.gilvum_PYR-GCK HGMRLLGFRPGRERPSTVYADPAGPAPEPR--------------------
Mvan_0717|M.vanbaalenii_PYR-1 HGMRLLGFRQRG--------------------------------------
MSMEG_0810|M.smegmatis_MC2_155 HGMRLLETLGGLGRRPAADQAPTPPTH-----------------------
MUL_4272|M.ulcerans_Agy99 LVLQTGAAGGPAAVESTDTLWLTRPL------------------------
MAV_1023|M.avium_104 APMLRRAARLPAAPSDRAGAPPTATADNQAK-------------------
MAB_2932c|M.abscessus_ATCC_199 LIGSFPDLGAEDEAGEMVAVDHTILQTLDEAELNNVVSLATAYREHFGFP
MMAR_4949|M.marinum_M LLNAFHDLGAEDETGRALAVDHVSLSNLDERDHNVVVELASAYREHFGFP
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0810|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4272|M.ulcerans_Agy99 --------------------------------------------------
MAV_1023|M.avium_104 --------------------------------------------------
MAB_2932c|M.abscessus_ATCC_199 LVVCARETERYERLLRSGWSRMENSATSERSFALIEIAKIANYRFDDLVA
MMAR_4949|M.marinum_M LIICARETERFDRVLKNGWSRMDNSPATERAYALIEAAKIANYRFSDLVA
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0810|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4272|M.ulcerans_Agy99 --------------------------------------------------
MAV_1023|M.avium_104 --------------------------------------------------
MAB_2932c|M.abscessus_ATCC_199 DANPITAARFGRFEDFHATR
MMAR_4949|M.marinum_M DANPIASARLSRLNELH---
Mflv_0188|M.gilvum_PYR-GCK --------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------
MSMEG_0810|M.smegmatis_MC2_155 --------------------
MUL_4272|M.ulcerans_Agy99 --------------------
MAV_1023|M.avium_104 --------------------