For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIFDWQVKSGDSVVAPHERLSWPRTTGIGLQHVVAMFGATFLVPVLTGFPPATTLLFSGLGTILFLIIT QNRLPSYLGSSFAIIAPTIAASTSHGPSGALGGIVIVGALLILIGAVVHFAGIGWIESLMPPVVTGSIVA LVGFNLAPAAKANFEEDAVTAAVVLVLLLLSLVVFRGLLGRLAIFTSVVIGYFVALARGKVDTSAIGAAD WVGLPEFTTPTFHLSVLPMFIPVVMVLVVENIGHVKSITAMTGIDNNSRVGRALAADGLATVLAGSGGGS ATTTYAENIGVMAATKVYSTAAYWIAGLAAILLGLSPKVGAVIASIPPGVLGGVTVALYGLVGILGVHIW VTNKVDFSKPINQFTAAIALVIGIADFTWKMGDLAFGGIALGAIGAVVVYHVMRMIGQWRGTVIIDGPAS DDAAAQDLTAGQKVAL
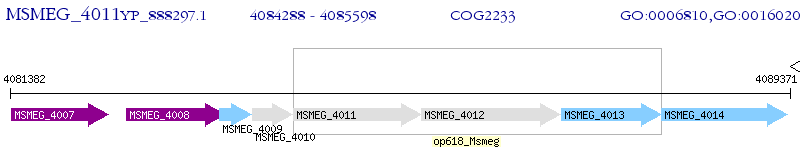
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4011 | - | - | 100% (436) | putative pyrimidine permease RutG |
| M. smegmatis MC2 155 | MSMEG_0810 | - | e-160 | 67.96% (412) | putative pyrimidine permease RutG |
| M. smegmatis MC2 155 | MSMEG_2570 | - | 3e-36 | 27.75% (418) | xanthine/uracil permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0188 | - | 1e-161 | 71.21% (396) | uracil-xanthine permease |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2932c | - | 9e-37 | 27.33% (472) | xanthine/uracil permease |
| M. marinum M | MMAR_4949 | - | 8e-38 | 29.07% (430) | xanthine/uracil permease |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0717 | - | 1e-160 | 69.95% (396) | uracil-xanthine permease |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0188|M.gilvum_PYR-GCK -MTLTWKRVDSTTDPDFVVAPDERLSWPSTLGLGAQHVVAMFGATFLVPV
Mvan_0717|M.vanbaalenii_PYR-1 --MRAWTKVDSRPDPDFVVAPDERLSWPRTLGLGAQHVVAMFGATFLVPV
MSMEG_4011|M.smegmatis_MC2_155 -MSIFDWQVKSG---DSVVAPHERLSWPRTTGIGLQHVVAMFGATFLVPV
MAB_2932c|M.abscessus_ATCC_199 MSADTTRPAAPGAQRKKVHPVDEVPSTGRLVTLGIQHVIAFYAGAVLVPL
MMAR_4949|M.marinum_M MSAPPTSPVR------RVHPVDEVLPIPKLATYGFQHVVAFYAGAVVVPI
. * . .* . * ***:*::..:.:**:
Mflv_0188|M.gilvum_PYR-GCK LTG-----------FPPATTLLFSGVGTILFLL---ITGNRLPSYLGSSF
Mvan_0717|M.vanbaalenii_PYR-1 LTG-----------FPPATTLLFSGVGTILFLL---ITGNRLPSYLGSSF
MSMEG_4011|M.smegmatis_MC2_155 LTG-----------FPPATTLLFSGLGTILFLI---ITQNRLPSYLGSSF
MAB_2932c|M.abscessus_ATCC_199 LIARAIDLDAEALTMLITADLFTCGVASILQAVGFWKIGVRLPLLQGVTF
MMAR_4949|M.marinum_M IIANAIDLPHQDLVKLITADLFTCGVASIIQSVGFWKIGVRLPLLQGVAF
: . :: *: .*:.:*: : *** * :*
Mflv_0188|M.gilvum_PYR-GCK SVIAPVTAATASHGTGSALGGIIAVGVLLIIIGAVVHVAGTRWIDITLPP
Mvan_0717|M.vanbaalenii_PYR-1 SVIAPVTAAVASEGTGSALGGIVAVGVLLILVGGVVHMVGTRWLDLTLPP
MSMEG_4011|M.smegmatis_MC2_155 AIIAPTIAASTSHGPSGALGGIVIVGALLILIGAVVHFAGIGWIESLMPP
MAB_2932c|M.abscessus_ATCC_199 ATLAPVIKIANDHGGGRVGLLTVYGAVIAAGAFTFLIAPFFARLIRFFPP
MMAR_4949|M.marinum_M AGVSPVIAIGLSHGGGTPSLLYVYGAVIVAGLFTILIAPIFTKLLRFFPP
: ::*. ..* . : ..: .: : :**
Mflv_0188|M.gilvum_PYR-GCK VVTGAIVALIGFNLAPAAKNNFEQGPLL---------------GFVTLVL
Mvan_0717|M.vanbaalenii_PYR-1 IVTGAIVALIGFNLAPAAKNNFEQGPLL---------------GLVTLVL
MSMEG_4011|M.smegmatis_MC2_155 VVTGSIVALVGFNLAPAAKANFEEDAVT---------------AAVVLVL
MAB_2932c|M.abscessus_ATCC_199 VVTGSVITIIGVSLLPVGIGDATRNPHSPVAAHDPGNGRWLMYALGTIGA
MMAR_4949|M.marinum_M VVTGTLITIIGLCLVPVGAGDAVTNPRT--GEHDPTNIRWLLYALGTIAI
:***::::::*. * *.. : .. . .:
Mflv_0188|M.gilvum_PYR-GCK LVVVLAAFRGLLGRLAIFVAVAAGYLLAVFLGEVDTSGIAAAAWIGLPD-
Mvan_0717|M.vanbaalenii_PYR-1 LVAALAFFRGMIGRLAIFVAVATGYVLALLLGEVDTSGIATAPWIGLPE-
MSMEG_4011|M.smegmatis_MC2_155 LLLSLVVFRGLLGRLAIFTSVVIGYFVALARGKVDTSAIGAADWVGLPE-
MAB_2932c|M.abscessus_ATCC_199 IVLMQRFFRGFWSTIAVLLGLVAGTVVAWLAGDTDFGRVGEADWIGFTSP
MMAR_4949|M.marinum_M IVAIQRVFSGFMATIAVLIGLVVGCVVAYGLGDMSFAKVDEASVIGFTEP
:: * *: . :*:: .:. * .:* *. . . : * :*:..
Mflv_0188|M.gilvum_PYR-GCK --FQAPTFTLSVLPLFLPAVVALVAENIGHVKSVGQMTKRDM-DPLTGRA
Mvan_0717|M.vanbaalenii_PYR-1 --FEAPTFTLSVLPMFLPAVIALIAENIGHVKSVSQMTKTDL-DPLMGRA
MSMEG_4011|M.smegmatis_MC2_155 --FTTPTFHLSVLPMFIPVVMVLVVENIGHVKSITAMTGIDN-NSRVGRA
MAB_2932c|M.abscessus_ATCC_199 FAFGAPRFDVIAVVSLIVVLMVTAVESTGAVFATAEIVGKRVRGADIAAT
MMAR_4949|M.marinum_M FVFGMPKFDFVACLTMIIVLMITSVESTGSTLATGEIVGKRIKAAQIGNV
* * * . . :: .:: .*. * . : :. . . .
Mflv_0188|M.gilvum_PYR-GCK LAADGVATTLAGFGGGSATTTYAENIGVMAATRVYSTAPYWIAALVAILL
Mvan_0717|M.vanbaalenii_PYR-1 LAADGVATTLAGFGGGSATTTYAENIGVMAATRVYSTAAYWVAALVAVAL
MSMEG_4011|M.smegmatis_MC2_155 LAADGLATVLAGSGGGSATTTYAENIGVMAATKVYSTAAYWIAGLAAILL
MAB_2932c|M.abscessus_ATCC_199 VRADGIATVIGGTFNSFPYTAFSENVGLVRLTGVKSRWVVAAAGVIMMVL
MMAR_4949|M.marinum_M LRADGLATVIGGIFNSFPYTAFSENVGLVRLTRVKSRWVVAAAGVVMIVL
: ***:**.:.* .. . *:::**:*:: * * * *.: : *
Mflv_0188|M.gilvum_PYR-GCK SLCPKVGAVIAAIPAGVLGGATVVLYGLVGIIGIRIWLTNHVDFSKPVNQ
Mvan_0717|M.vanbaalenii_PYR-1 SLCPKVGAVIVAVPAGVLGGATVVLYGLVGILGVRIWLTNHVDFSKPINQ
MSMEG_4011|M.smegmatis_MC2_155 GLSPKVGAVIASIPPGVLGGVTVALYGLVGILGVHIWVTNKVDFSKPINQ
MAB_2932c|M.abscessus_ATCC_199 GVLPKTAKIVESIPAPVLGGAAMTLFATVAVVGIQTLG--KVDFNDHRNL
MMAR_4949|M.marinum_M GFLPKVAAVVSAVPSPVLGGAALTLFATVAVVGIQTLG--KVDFTDHRNL
.. **.. :: ::*. ****.::.*:. *.::*:: :***.. *
Mflv_0188|M.gilvum_PYR-GCK MTAAIPLIIG----------------------------IADFTWSAGSLT
Mvan_0717|M.vanbaalenii_PYR-1 MTAAIPLIIG----------------------------IADFTWSAGGLT
MSMEG_4011|M.smegmatis_MC2_155 FTAAIALVIG----------------------------IADFTWKMGDLA
MAB_2932c|M.abscessus_ATCC_199 IIASTSLAMGMYVQFSQSSGPATVLDHGRGIAVPALPGIDQAVPGLLQIP
MMAR_4949|M.marinum_M IIVTTSLALALWVTS--------------------YPDVAESMPSGVDLI
: .: .* :. : : :
Mflv_0188|M.gilvum_PYR-GCK FT-GIALGSIAALLIYHGMRLLGFRPGRERPSTVYADPAGPAPEPR----
Mvan_0717|M.vanbaalenii_PYR-1 FT-GIALGSIAALLIYHGMRLLGFRQRG----------------------
MSMEG_4011|M.smegmatis_MC2_155 FG-GIALGAIGAVVVYHVMRMIGQWRGTVIIDGPASDDAAAQDLTAGQKV
MAB_2932c|M.abscessus_ATCC_199 FSTGITMGAITAIVLNLLFFHTGSRGVAVAGGGAIRLAQVNEMTLEQFRR
MMAR_4949|M.marinum_M FGSGISIGAVAAIVLNIVFFHTGSHGPRVAGGGSITLHEVNAMTRERFTE
* **::*:: *::: : *
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4011|M.smegmatis_MC2_155 AL------------------------------------------------
MAB_2932c|M.abscessus_ATCC_199 TFGGLVQDVHWVVDRAYRQRPFADAHDLRSAFQEAMLTGSDEEQLELIGS
MMAR_4949|M.marinum_M VFGHVVQDVPWVIDRAYEHRPFADTAALREAFQDAVLTGSSEQQMELLNA
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4011|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2932c|M.abscessus_ATCC_199 FPDLGAEDEAGEMVAVDHTILQTLDEAELNNVVSLATAYREHFGFPLVVC
MMAR_4949|M.marinum_M FHDLGAEDETGRALAVDHVSLSNLDERDHNVVVELASAYREHFGFPLIIC
Mflv_0188|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0717|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4011|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2932c|M.abscessus_ATCC_199 ARETERYERLLRSGWSRMENSATSERSFALIEIAKIANYRFDDLVADANP
MMAR_4949|M.marinum_M ARETERFDRVLKNGWSRMDNSPATERAYALIEAAKIANYRFSDLVADANP
Mflv_0188|M.gilvum_PYR-GCK ----------------
Mvan_0717|M.vanbaalenii_PYR-1 ----------------
MSMEG_4011|M.smegmatis_MC2_155 ----------------
MAB_2932c|M.abscessus_ATCC_199 ITAARFGRFEDFHATR
MMAR_4949|M.marinum_M IASARLSRLNELH---