For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEIIDPSAPGKAPSGKSSEAIQRLKPNAVGLVGVMFMAVATAAPITAMVGNVPIAVGFGNGAYAPAGYF VATIVLTLFAIGYAAMSKHITATGAFYGYISHGLGRIVGLGAGFLTALAYMVFEASLIGIFSFFGNDLFK SFFDVDVPWIVFAVVMLAVNTVLTYFDINLAAKVLGVFLITEIVMLALMALSVVFTGGGPQGWSLGSLNP LNGFQSLTGEVAGTDGSMIAVAGSAGVGLFFAFWSWVGFESSAMYGEESRNPKKIIPIAVVSSVIGIGAF YVIVSWLAIVGTGPQNAIALAQDSETAGNIFFNPVHEHLGTWAVDLFKILLMTGSFACGMAFHNCAARYL YAIGRENVVPGMRKTIGATHPVHGSPYIAGFVQTAFAAVVVLFFDLSGRDPYTELYGLMALLGTTAIMIV QALAAFSVISYFHVHKRHPETAHWFKTFLAPLLGGTGMLYVIYLLAKNASFAAGDAATDWIFAAIPYVVG IVGIGGVLLALFLKFKDPNRYAELGRTVLQESHER
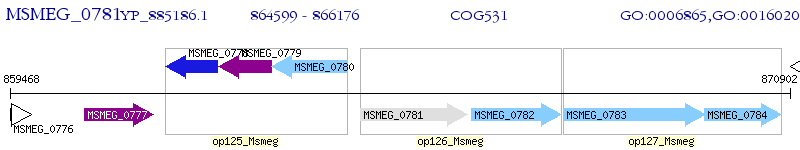
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0781 | - | - | 100% (525) | amino acid permease |
| M. smegmatis MC2 155 | MSMEG_0279 | - | 0.0 | 63.62% (514) | amino acid permease |
| M. smegmatis MC2 155 | MSMEG_3821 | - | e-135 | 47.69% (497) | amino acid permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2347c | rocE | 1e-16 | 22.07% (435) | cationic amino acid transport integral membrane protein RocE |
| M. gilvum PYR-GCK | Mflv_1244 | - | 0.0 | 60.72% (527) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2320c | rocE | 1e-16 | 22.07% (435) | cationic amino acid transport integral membrane protein RocE |
| M. leprae Br4923 | MLBr_01974 | - | 7e-13 | 22.83% (381) | putative permease |
| M. abscessus ATCC 19977 | MAB_0950c | - | 0.0 | 77.80% (527) | putative amino acid permease family protein |
| M. marinum M | MMAR_0869 | - | 2e-21 | 24.30% (465) | amino acid permease |
| M. avium 104 | MAV_2087 | - | 5e-14 | 21.63% (453) | amino acid transporter |
| M. thermoresistible (build 8) | TH_4382 | - | 6e-15 | 24.10% (498) | PUTATIVE amino acid permease-associated region |
| M. ulcerans Agy99 | MUL_1244 | rocE | 3e-16 | 22.58% (434) | cationic amino acid transport integral membrane protein RocE |
| M. vanbaalenii PYR-1 | Mvan_5564 | - | 1e-180 | 60.78% (510) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2347c|M.bovis_AF2122/97 ---------------------------------------------MPTTS
Rv2320c|M.tuberculosis_H37Rv ---------------------------------------------MPTTS
MUL_1244|M.ulcerans_Agy99 ---------------------------------------------MPATS
MAV_2087|M.avium_104 --------------------------------------------------
MSMEG_0781|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0950c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1244|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5564|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0869|M.marinum_M --------------------------------------------------
TH_4382|M.thermoresistible__bu --------------------------------------------------
MLBr_01974|M.leprae_Br4923 MFLSADVSCIEIVDINCSELRNDRRIPPQLALFCTNAANKAESAVTSHGQ
Mb2347c|M.bovis_AF2122/97 MSLRELMLRRRPVSG-APVASGASGNLKR---------------------
Rv2320c|M.tuberculosis_H37Rv MSLRELMLRRRPVSG-APVASGASGNLKR---------------------
MUL_1244|M.ulcerans_Agy99 IRLREQMLRRRPAGG-APVAEGAAGSLKR---------------------
MAV_2087|M.avium_104 ------------------MAPGTADHLRR---------------------
MSMEG_0781|M.smegmatis_MC2_155 --MSEIIDPSAPGKAPSGKSSEAIQRLKPN--------------------
MAB_0950c|M.abscessus_ATCC_199 --MSDTIDPPVG----TTTEPDSMQRLKPN--------------------
Mflv_1244|M.gilvum_PYR-GCK --MTESAERTAPG---QTTPTASVQRLKRN--------------------
Mvan_5564|M.vanbaalenii_PYR-1 -----MTEDVVPR---QTAPTDSVQRLKRN--------------------
MMAR_0869|M.marinum_M ----------------MAVTTGARSELQK---------------------
TH_4382|M.thermoresistible__bu ---------------MSGTRTEQAQPELRR--------------------
MLBr_01974|M.leprae_Br4923 LPIMANRRWAPHTRRQSGRATSTCSSLMRFSASESRAHSRFECYTLVRWS
Mb2347c|M.bovis_AF2122/97 ----------SFGTFQLTMFGVG--ATIGTGIFFVLAQAVPEAGPGVIVS
Rv2320c|M.tuberculosis_H37Rv ----------SFGTFQLTMFGVG--ATIGTGIFFVLAQAVPEAGPGVIVS
MUL_1244|M.ulcerans_Agy99 ----------SFGTFELTMFGVG--STVGTGIFFVLAEAVPEAGPAVIVS
MAV_2087|M.avium_104 ----------GIGTFQLTMFGVG--STIGTGIFFVMSQAVPEAGPAVIVS
MSMEG_0781|M.smegmatis_MC2_155 ----------AVGLVGVMFMAVATAAPITAMVGNVPIAVGFGNGAYAPAG
MAB_0950c|M.abscessus_ATCC_199 ----------AVGLIGVLFMAVATAAPITAMVGNVPMAVGFGNGAYAPAG
Mflv_1244|M.gilvum_PYR-GCK ----------AVGTFGVIFMAVATAAPITAMVGNVPIAVGFGNGSHAPAG
Mvan_5564|M.vanbaalenii_PYR-1 ----------AVGTFGVIFMAVATAAPITAMVGNVPIAVGFGNGSHAPAG
MMAR_0869|M.marinum_M ----------ALGQRSAVLFGLAYMTPIIVLG--IFGVIAERSKGASAGS
TH_4382|M.thermoresistible__bu ----------VLGPGLLLLFIVG--DILGTGIYALVGDVAGEVGGAAWLP
MLBr_01974|M.leprae_Br4923 IRGQGGRAISKLGFWSVVMLGIN--SIIGAGIFMTPGEVIKIAGPFAPIA
.* :: : : .
Mb2347c|M.bovis_AF2122/97 FIIAGIAAGLAAICYAELASAVPISGSAYSYAYTTLGEAVAMVVAACLLL
Rv2320c|M.tuberculosis_H37Rv FIIAGIAAGLAAICYAELASAVPISGSAYSYAYTTLGEAVAMVVAACLLL
MUL_1244|M.ulcerans_Agy99 FIIAGVAAGLAAICCAELASAVPVSGSSYSYAYATLGEAVAMGVASCLLL
MAV_2087|M.avium_104 FLLAGVAAGLAAVCYAELASAVPVSGSSYSYAYTTLGEVVAMGVAACLLL
MSMEG_0781|M.smegmatis_MC2_155 YFVATIVLTLFAIGYAAMSKHITATGAFYGYISHGLGRIVGLGAGFLTAL
MAB_0950c|M.abscessus_ATCC_199 YLVATIVLTLFAIGYAAMSRHIVATGAFYGYISHGLGRVVGLGSGFLITL
Mflv_1244|M.gilvum_PYR-GCK YLVATIVLGLFAIGYATMAKHITATGAFYGYISHGLGRVMGMASGLIITL
Mvan_5564|M.vanbaalenii_PYR-1 YLVATVVLGLFAIGYATMAKHITATGAFYGYISHGLGRVMGMASGGIITM
MMAR_0869|M.marinum_M YLLATVAMLFTAQSYGVMARHFPVAGSAYTYVRKALDARVGFMVGWAVLL
TH_4382|M.thermoresistible__bu FLIAFLIATVTAFSYLELVTKYPQAAGAALYVHKAFGIQFVTFLVAFIVM
MLBr_01974|M.leprae_Br4923 YTLAGIFAAVLAIVFATAAKYVKTNGSSYAYTTAAFGHRIGIYVGVTHAI
: :* : . * .. * :. . :
Mb2347c|M.bovis_AF2122/97 EYGVATAAVAVGWS------GYVNKLLSNLFGFQMPHVLSAAPWDTHPG-
Rv2320c|M.tuberculosis_H37Rv EYGVATAAVAVGWS------GYVNKLLSNLFGFQMPHVLSAAPWDTHPG-
MUL_1244|M.ulcerans_Agy99 EYGVATAAVAVGLS------GYVNKLLHNVFGFQLPQALSEAPWDERPGE
MAV_2087|M.avium_104 EYGVATAAVSVNWS------GYLNKLLSNVVGFQLPHALSAAPWDAQPG-
MSMEG_0781|M.smegmatis_MC2_155 AYMVFEASLIGIFS------FFGNDLFKSFFDVDVPWIVFAVVMLAVNT-
MAB_0950c|M.abscessus_ATCC_199 AYMVFEASLVGIFS------FFANDLFQTFFQVDVPWVVFAVAMLATNA-
Mflv_1244|M.gilvum_PYR-GCK AYIVFEASLIGIFA------FFFQNFLSSQLGITIHWLVPALLMLVLNS-
Mvan_5564|M.vanbaalenii_PYR-1 AYVVFEASLIGIFA------FFFQNFLSSQFGFDIHWLIPALLMLALNC-
MMAR_0869|M.marinum_M DYLFLPLVIWLIGG------SYLQDRFPTVP--FWTWIVGFAALTTVLN-
TH_4382|M.thermoresistible__bu CSGITSASTAS---------RFFAANLEVGLGQEWGRLGVAGVALLFMAL
MLBr_01974|M.leprae_Br4923 TASIAWGVLASSFVSTLLRVAFPEKIWADNDELISVKTLTFLVFIAVLLA
. :
Mb2347c|M.bovis_AF2122/97 ---WVNLPAVILIGLCALLLIRGASESARVNAIMVLIKLGVLGMFMIIAF
Rv2320c|M.tuberculosis_H37Rv ---WVNLPAVILIGLCALLLIRGASESARVNAIMVLIKLGVLGMFMIIAF
MUL_1244|M.ulcerans_Agy99 AAGYLNLPAIILIGLCALLLIRGVSESAKVNAIMVLIKLGVLGMFVILAL
MAV_2087|M.avium_104 ---YVNLPAVMLIGMCALLLIRGASESAKVNAIMVMVKLGVLVVFGILAF
MSMEG_0781|M.smegmatis_MC2_155 --VLTYFDINLAAKVLGVFLITEIVMLALMALSVVFTGGGPQGWSLGS-L
MAB_0950c|M.abscessus_ATCC_199 --VLTYFDLNLAARVLGVFLVTEILMLSLLALSVLFAGGGPQGWSWGS-L
Mflv_1244|M.gilvum_PYR-GCK --ILTYFDVNLTAKVLGVFLITEIFMLALGAVAVAVKGGGPDGFAIAETL
Mvan_5564|M.vanbaalenii_PYR-1 --ILTYFDVNLTAKVLGVFLVTEIVMLSLGAIAVAVKGGGPDGFAVGEIL
MMAR_0869|M.marinum_M -------LIGLKVADRANFILMTFQLLILAFFVALTIGYLVGDSRSLLSL
TH_4382|M.thermoresistible__bu LAMVNLRGVGESVRLNVLLTIVEISGLLLVIFVGLWAFTGGGADVDFTRV
MLBr_01974|M.leprae_Br4923 INIFGNLVIRWANGISALGKIFALSVFIAGGLWTIITQHVNNYVTSRSAY
:
Mb2347c|M.bovis_AF2122/97 SAYSADHLK-------DFVPFGVAG--IGSAAGTIFFSYIGLDAVSTAGD
Rv2320c|M.tuberculosis_H37Rv SAYSADHLK-------DFVPFGVAG--IGSAAGTIFFSYIGLDAVSTAGD
MUL_1244|M.ulcerans_Agy99 TAYDTNHLK-------DFAPFGVAG--VGAAAGTIFFSFIGLDAVSTAGE
MAV_2087|M.avium_104 TAFDMHHLD-------DFAPFGVAG--VGTAAGTIFFSYIGLDAVSTAGD
MSMEG_0781|M.smegmatis_MC2_155 NPLNGFQSLTGEVAGTDGSMIAVAG-SAGVGLFFAFWSWVGFESSAMYGE
MAB_0950c|M.abscessus_ATCC_199 NPLNGFHDLGGVVQGPAG-PLAVAG-SAGVGLFFAFWSWVGFESSAMYAE
Mflv_1244|M.gilvum_PYR-GCK NPVGAFQ------------PAAIAGASAGLGLFFAFWSWVGFESTAMYGE
Mvan_5564|M.vanbaalenii_PYR-1 NPVGAFQ------------PAAIAGASAGLGLFFAFWSWVGFESTAMYGE
MMAR_0869|M.marinum_M TPFTGNG--------------GFTA--ITAGAAVAAYSFLGFDAVSTLTE
TH_4382|M.thermoresistible__bu VAFETTTDK-------------SVFLAVSAATSLAFFAMVGFEDSVNMAE
MLBr_01974|M.leprae_Br4923 QPTTYSLLG-------VTEIGKSTAASMTLATIVALYAFTGFESIANAAE
. . . :: *:: :
Mb2347c|M.bovis_AF2122/97 EVKDPQKTMPRALIAALVVVTGVYVLVALAALG-TQPWQDFAEQET----
Rv2320c|M.tuberculosis_H37Rv EVKDPQKTMPRALIAALVVVTGVYVLVALAALG-TQPWQDFAEQET----
MUL_1244|M.ulcerans_Agy99 EVRNPQQSVPRALIAALFVVTGVYVLVAFAALG-TQPWQMFAGQEE----
MAV_2087|M.avium_104 EVTNPQKTMPRALIAALLTVTGVYVFVALAALG-TQPWQDFGGQQE----
MSMEG_0781|M.smegmatis_MC2_155 ESRNPKKIIPIAVVSSVIGIGAFYVIVSWLAIVGTGPQNAIALAQDSETA
MAB_0950c|M.abscessus_ATCC_199 ESKNPKKIIPIAIVTSVVGVGLFYVLVSWLAIVGTGPENAVALAQDSSTA
Mflv_1244|M.gilvum_PYR-GCK ESKDPKRIIPRATMIAVLGVGIFYVFVSWMAIAGTGPQQAVELAQSADTS
Mvan_5564|M.vanbaalenii_PYR-1 ESKDPKRIIPRATMIAVLGVGLFYVFVSWMAIAGTGPKQAVELAQSADTS
MMAR_0869|M.marinum_M ETHDAERTIPRAIVLIALIGGAIFVTVAYVVSL-VAPGSSFPDADS----
TH_4382|M.thermoresistible__bu ETRDPVVIFPRALLTGLGIAGAVYVVVAIVAVALVPVGTLQASDVP----
MLBr_01974|M.leprae_Br4923 EMNTPDRTLPKAIPLTILTVGVIYILALVVAML-LGSDKIVATNHT----
* . .* * .:: . .
Mb2347c|M.bovis_AF2122/97 AGLAIILDNVTHGEWASTILAAGAVVSIFTVTLVTMYGQTRILFAMGRDG
Rv2320c|M.tuberculosis_H37Rv AGLAIILDNVTHGEWASTILAAGAVVSIFTVTLVTMYGQTRILFAMGRDG
MUL_1244|M.ulcerans_Agy99 AGLATILDNVTHSAWASTVLAMGAVISIFTVTLVTMYGQTRILFAMGRDG
MAV_2087|M.avium_104 AGLATILDHVTHGSWASTILAAGAVISIFSVTLVTMYGITRILFAMGRDG
MSMEG_0781|M.smegmatis_MC2_155 GNIFFNPVHEHLGTWAVDLFKILLMTGSFACGMAFHNCAARYLYAIGREN
MAB_0950c|M.abscessus_ATCC_199 GNIFFGPVQSHLGVWAVNLFKVLLMTGSFACGMAFHNCAARYLYALGREN
Mflv_1244|M.gilvum_PYR-GCK SEIFFAPVRDTYGEWAITLFNILLVTGSFACGMAFHNCASRYLYALGREG
Mvan_5564|M.vanbaalenii_PYR-1 SEIFFAPVRTTYGEWAITLFNILLVSGSFACGMAFHNCASRYLYALGREG
MMAR_0869|M.marinum_M --LSEGVARTIGGNLFAAVFLAALIIGQFTSGLAAQAAVSRLLYAMGRDG
TH_4382|M.thermoresistible__bu --LVEVVKAGAPGLPIEELLPWISMFAVSNTALINMLMASRLIYGMARQR
MLBr_01974|M.leprae_Br4923 ----VKLAAAVGNNTFRTIIVVGALVSMFGINVAASFGAPRLWTALSDGR
. :: : . : .* .:.
Mb2347c|M.bovis_AF2122/97 LLP---ARFAKVNPRTMTPVHNTVIVAIFASTLAAFIPLDSL--ADMVSI
Rv2320c|M.tuberculosis_H37Rv LLP---ARFAKVNPRTMTPVHNTVIVAIFASTLAAFIPLDSL--ADMVSI
MUL_1244|M.ulcerans_Agy99 LLP---ARFAAVHPRFQTPVSNTVVVAIAAAVLAALVPLNRL--ADMVSI
MAV_2087|M.avium_104 LLP---PRFARVNPRTMTPVNNTVIVAVAAATLAAFIPLQNL--ADMVSI
MSMEG_0781|M.smegmatis_MC2_155 VVPGMRKTIGATHPVHGSPYIAGFVQTAFAAVVVLFFDLSGR--DPYTEL
MAB_0950c|M.abscessus_ATCC_199 VIPGMRKTIGSTHSTHGSPHVAGFVQTAFATVVVLFFAATGR--DPYTGL
Mflv_1244|M.gilvum_PYR-GCK LSTGLQKTLGATHKTHGSPYIASFVQSGIALVLVLAFFFAGM--DPYIHM
Mvan_5564|M.vanbaalenii_PYR-1 LSTRLQDTLGATHKTHGSPHIASFVQTGITLVLILAFFAAGM--DPYIHM
MMAR_0869|M.marinum_M VLP--RRVFGVVSAKFHTPMFN-IMLTGAVGMAAILLSVATS--TSFINF
TH_4382|M.thermoresistible__bu VLP---PVLGSVHPRRRTPWVAIIFTTLIAFGLIFYVTAFAD--STAIAV
MLBr_01974|M.leprae_Br4923 ILP----TRLSSKNRFGVPIVAFGITAFLTLAFPLSLRFDSLNLTGLAVI
: . * . : . . .
Mb2347c|M.bovis_AF2122/97 GT-------LTAFSVVAVGVIVLRVR-------------EPDLPRGFKVP
Rv2320c|M.tuberculosis_H37Rv GT-------LTAFSVVAVGVIVLRVR-------------EPDLPRGFKVP
MUL_1244|M.ulcerans_Agy99 GT-------LTAFIVVSVGVIILRVR-------------EPALPRGFWVP
MAV_2087|M.avium_104 GT-------LTAFVVVSVGVIVLRVR-------------EPDLPRGFRVP
MSMEG_0781|M.smegmatis_MC2_155 YG-------LMALLGTTAIMIVQALAAFSVISYFHVHKRHPETAHWFKTF
MAB_0950c|M.abscessus_ATCC_199 YG-------LMALLGTTAILIVQALTAFAVISYFHVLRHHPETAHWFRTL
Mflv_1244|M.gilvum_PYR-GCK YT-------LLAILGTMAILIVQSLCAFSVVAYFHFHKNHPSSAHWFKTF
Mvan_5564|M.vanbaalenii_PYR-1 YT-------LLAILGTMAILIVQSLCAFSVIAYFHFHKNHPSSAHWFKTF
MMAR_0869|M.marinum_M GA-------FTAFTLVNVSVVVYFVR---------------ERGAGLPRW
TH_4382|M.thermoresistible__bu LGGTTSLLLLAVFAMVNVAVLVLRRD-----------ESRDPVVARRHFR
MLBr_01974|M.leprae_Br4923 AR-------FIQFIIVPIALFALARS-----------RTTNSVYVKRNVF
: : . :.
Mb2347c|M.bovis_AF2122/97 GYPVTPVLSVLACGYILAS-----------------LHWYTWLAFSGWVA
Rv2320c|M.tuberculosis_H37Rv GYPVTPVLSVLACGYILAS-----------------LHWYTWLAFSGWVA
MUL_1244|M.ulcerans_Agy99 GYPVTPILSILACGYLLYS-----------------LHWYTWIAFGGWVG
MAV_2087|M.avium_104 GYPVTPVLSIMACGYILAS-----------------LHWYTWIAFSGWVL
MSMEG_0781|M.smegmatis_MC2_155 LAPLLGGTGMLYVIYLLAKNASFAAGDAATDWIFAAIPYVVGIVGIGGVL
MAB_0950c|M.abscessus_ATCC_199 VAPLLGGLGMVYVVYLLAKHASFAAGTASNDWVFTAIPWVVAAAGIGGIA
Mflv_1244|M.gilvum_PYR-GCK LAPLTGGIGMLYVVYLLWEHKESAAGAASGTLLFKLTPWIVVGVFAFGAA
Mvan_5564|M.vanbaalenii_PYR-1 LAPLLGGIGMLYVVYLLWEHKESAAGAASGTLLFKLTPWIVVGLFALGAA
MMAR_0869|M.marinum_M RYLVLPVIGAAVDIYLLAH-----------------LASDALIVGLSWAA
TH_4382|M.thermoresistible__bu TPTFLPVVGFVASLYLVTPLSGRP-----------AQQYLLSVVLIVIGV
MLBr_01974|M.leprae_Br4923 TDKVLPITAVVVSAGLMVS------------------FDYRSIFLTRGGP
. . ::
Mb2347c|M.bovis_AF2122/97 VAVIFYLMWGRHHSALNEEVP----------
Rv2320c|M.tuberculosis_H37Rv VAVIFYLMWGRHHSALNEEVP----------
MUL_1244|M.ulcerans_Agy99 IALIFYLFWGRHHSRLNAPALHDNPTGEER-
MAV_2087|M.avium_104 LALIFYFVWGRHHSALNDAAVD--PSGQER-
MSMEG_0781|M.smegmatis_MC2_155 LALFLKFKDPNRYAELGRTVLQESHER----
MAB_0950c|M.abscessus_ATCC_199 LALTLKYTAPHRYDDLGRTVLEEAHER----
Mflv_1244|M.gilvum_PYR-GCK MALYFKFKDKRRYELIGRIVYDDSEVRD---
Mvan_5564|M.vanbaalenii_PYR-1 MALYFKFNDKRRYELIGRIVYDDSEIRD---
MMAR_0869|M.marinum_M IGFLYLLWLTRGLTRQPPDLAGAAERG----
TH_4382|M.thermoresistible__bu LLFGVTMAINRQLGIPTRAVGDPAKMVLPPD
MLBr_01974|M.leprae_Br4923 NYFSISLIAITFIGIPAVAYLHYYRTSGIK-
.