For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VYYVGPVTAGAAPRILKHQVVRAELDRMLDGMRIGDPFPAEREIAEQFEVARETVRQALRELLIDGRVER RGRTTVVARPKIRQPLGMGSYTEAAKAQGLSAGRILVAWSDLTADEVLAGVLGVDVGAPVLQLERVLTTD GVRVGLETTKLPAQRYPGLRETFDHEASLYAEIRSRGIAFTRTVDTIDTALPDAREAALLGADARTPMFL LNRVSYDQDDVAIEQRRSLYRGDRMTFTAVMHAKNSAIVS
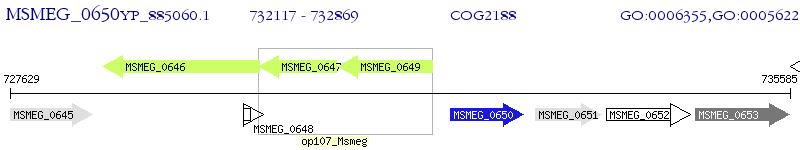
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0650 | - | - | 100% (250) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0286 | - | 1e-56 | 53.39% (221) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0268 | - | 3e-18 | 28.33% (233) | transcriptional regulator, GntR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0816c | - | 2e-08 | 26.18% (233) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2652 | - | 1e-23 | 36.32% (201) | regulatory protein GntR, HTH |
| M. tuberculosis H37Rv | Rv0792c | - | 2e-08 | 26.18% (233) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1498c | - | 3e-61 | 51.75% (228) | GntR family transcriptional regulator |
| M. marinum M | MMAR_4842 | - | 4e-12 | 25.85% (205) | regulatory protein |
| M. avium 104 | MAV_5032 | - | 1e-12 | 27.60% (221) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_3932 | - | 2e-15 | 28.88% (232) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_0525 | - | 6e-11 | 25.96% (208) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0056 | - | 2e-62 | 53.78% (225) | UbiC transcription regulator-associated domain-containing |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1498c|M.abscessus_ATCC_199 --------------MTADVRLPKHYQVRTELEHILSA--LNEGDPVPPER
Mvan_0056|M.vanbaalenii_PYR-1 ------------------MNVPKPYLVRTGLDGILDG--LQEGDPFPPER
MSMEG_0650|M.smegmatis_MC2_155 ------MYYVGPVTAGAAPRILKHQVVRAELDRMLDG--MRIGDPFPAER
Mb0816c|M.bovis_AF2122/97 -MTSVKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLPSER
Rv0792c|M.tuberculosis_H37Rv -MTSVKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLPSER
MMAR_4842|M.marinum_M MEGCRVLQLGQIDRTDDK---PPYRQIASMLREAISSSRFPPGERLPSEA
MUL_0525|M.ulcerans_Agy99 MEGSQVLQLGQIDRADDK---PPYRQIASMLRGAIISGELEAGGRLPSEA
Mflv_2652|M.gilvum_PYR-GCK --MSTTEADLRGYSAKMPAGTPLYMSIASDVRDLIASEALGPHTLLPSER
TH_3932|M.thermoresistible__bu --MTRTATPMRSTTINRFADRPLYRQLSEVLEARLIER-ARPGDRLPSEA
MAV_5032|M.avium_104 -------------------------MLRSQILEGAYGGLAAARPALPSES
: : .*.*
MAB_1498c|M.abscessus_ATCC_199 DLAERFAVSRETVRQALHELLVEGRIERR-GRGTVVAAP-----------
Mvan_0056|M.vanbaalenii_PYR-1 ELAARFGVARETVRQALHELLVEGRIERR-GRGTVVSRP-----------
MSMEG_0650|M.smegmatis_MC2_155 EIAEQFEVARETVRQALRELLIDGRVERR-GRTTVVARP-----------
Mb0816c|M.bovis_AF2122/97 ELIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP-----------
Rv0792c|M.tuberculosis_H37Rv ELIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP-----------
MMAR_4842|M.marinum_M ELIEHFGVARMTVRQAMQELRSDGLVISQHGRGVFVRPTPPIRRLASDRF
MUL_0525|M.ulcerans_Agy99 ELIEYFGVARMTVRQAVQELRTEGLVISEHGRGVFVRPAPPIRWLASDRF
Mflv_2652|M.gilvum_PYR-GCK ELAEQHGVSRMTARQALSLLESEGVVYRKPPRGTFVAEP-----------
TH_3932|M.thermoresistible__bu ELSGQFGVNRLTVRRALHELAQRGIIETVHGRGSFVARQP---------V
MAV_5032|M.avium_104 ALAAELGVSRNAIREALELLRGEGLITRVQGSGTFVTGAK----------
: : * : * *: * * : .*
MAB_1498c|M.abscessus_ATCC_199 -KLLQPLSLRSYTEGAQSQGRTPGRLLVTWENITGDADLCRDLGIRGGST
Mvan_0056|M.vanbaalenii_PYR-1 -KLVQPLSLRSYTEGAQRMGRTPGRLLVTWENITVDAEIAAALDVSPSAR
MSMEG_0650|M.smegmatis_MC2_155 -KIRQPLGMGSYTEAAKAQGLSAGRILVAWSDLTADEVLAGVLGVDVGAP
Mb0816c|M.bovis_AF2122/97 VEQELSCGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGL---VE
Rv0792c|M.tuberculosis_H37Rv VEQELSCGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGL---VE
MMAR_4842|M.marinum_M ARQHRAEGKAAFSVEAEKSGYSPQVDNIAVSREKPNSFIAEQLRISADDD
MUL_0525|M.ulcerans_Agy99 ARQHRANGKAAFTVEAERSGHTPQVDNITVSREKPDSVVAERLRLTADDE
Mflv_2652|M.gilvum_PYR-GCK ---RVRFHIGSFSEEVARMGRRPAAQLLWAEHQAATPAVRRALGLDEGAA
TH_3932|M.thermoresistible__bu RYRLSATRDASFTRGMRELGHPVRIEVLGAETTRCR---RLRTELHTAGE
MAV_5032|M.avium_104 -LRQDLNRLKGLAESLAGHRLPVENKVLSIRESTATPFVAAKLQLPENAP
: : :
MAB_1498c|M.abscessus_ATCC_199 VIHLERVLLADGAKLGLESTYLAKARFAELRNTFDPSTS---LYAAIRAL
Mvan_0056|M.vanbaalenii_PYR-1 VMHLERVLLADDVRIGLESTYLPRHRFGDLEQHFDPTTS---LYSAIRAR
MSMEG_0650|M.smegmatis_MC2_155 VLQLERVLTTDGVRVGLETTKLPAQRYPGLRETFDHEAS---LYAEIRSR
Mb0816c|M.bovis_AF2122/97 VLCIRRRIRTGDQPLALVTAYLPPGVGPAVEPLLSGSADTETTYAMWERR
Rv0792c|M.tuberculosis_H37Rv VLCIRRRIRTGDQPLALVTAYLPPGVGPAVEPLLSGSADTETTYAMWERR
MMAR_4842|M.marinum_M IVVRSRRYLANGRPVETAVSYIPAAFAEGTR-IEQVDTGPGGIYARLEEH
MUL_0525|M.ulcerans_Agy99 VVVRSRRYLADGKPVETATSYIPVAFAQGTK-IEQTDTGPGGIYARFEEA
Mflv_2652|M.gilvum_PYR-GCK VHVFHRLRSVDEVPFALETTFLPAELTPGILDMTEDGSL----WAVLRSK
TH_3932|M.thermoresistible__bu VLTTTTLRHVDGQPWSVSVTSIALDRFPGIAQQWSGSTS---LFDFLWQT
MAV_5032|M.avium_104 ILFIERLRSVGGLPLSLDTTSLRIG---AIRIDADLDKQ--DVFALIEQE
: .. : : . :
MAB_1498c|M.abscessus_ATCC_199 G-VQFGSAVERIETVLASPREASLLDTSIAMPMLLMNRRTLDTDGRPIER
Mvan_0056|M.vanbaalenii_PYR-1 G-VKFGSATERIETVLPSPREAELLESTTAMPMLLLNRRSVDVDGVPIEV
MSMEG_0650|M.smegmatis_MC2_155 G-IAFTRTVDTIDTALPDAREAALLGADARTPMFLLNRVSYDQDDVAIEQ
Mb0816c|M.bovis_AF2122/97 LGVRIAQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEV
Rv0792c|M.tuberculosis_H37Rv LGVRIAQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEV
MMAR_4842|M.marinum_M G-HVLARFTEEVAARMPTPEERRQLELDSGVPVLTVLRTAYDTNDVAVEV
MUL_0525|M.ulcerans_Agy99 G-HTLARFNEEVGARMPTPEERRALQLPPGAPVLTVVRTAYDTKDVAVEV
Mflv_2652|M.gilvum_PYR-GCK YAIELARTTAVLESIVLDDASSVQLGVRAGSAGTLLTRTTYDSADRCVEY
TH_3932|M.thermoresistible__bu FGVRMRRAYRSFSAVLADLAEAQHLGLRTGAPVLEMRGLNVDQHGAPVAV
MAV_5032|M.avium_104 LGERLGWAEITVESVLADPNTATLLQIRTGAPLLLLHRLTHLRDGTPFDL
: . : * . : . .
MAB_1498c|M.abscessus_ATCC_199 VRSLFRGDRVAFEAVLKE----------------
Mvan_0056|M.vanbaalenii_PYR-1 VRALYRGDRVAFEATLDDR---------------
MSMEG_0650|M.smegmatis_MC2_155 RRSLYRGDRMTFTAVMHAKNSAIVS---------
Mb0816c|M.bovis_AF2122/97 VVFHHRPERYQFSVTLPRTLPGSGAGIIEKRDFA
Rv0792c|M.tuberculosis_H37Rv VVFHHRPERYQFSVTLPRTLPGSGAGIIEKRDFA
MMAR_4842|M.marinum_M CDTVKVASAYLLEYEFPAR---------------
MUL_0525|M.ulcerans_Agy99 CDTVKVASAYLLEYDFSAR---------------
Mflv_2652|M.gilvum_PYR-GCK ARDVYRADRAAFEVSEELRSQRLSAV--------
TH_3932|M.thermoresistible__bu VRHRFRGDRVELTVDLA-----------------
MAV_5032|M.avium_104 ETVRYRGDRCSLVTTATREDAPPQ----------
. :