For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTIHTSQRTPAAERIAVRCVDSDVHPVPRRGVLGEYIPEPWRSSYFGTHDVGELIYYDAPDYAHAYAMRV DTFPSDGEFAGSDPDLAFRQLIMEAGADIAILEPAAYSAHIPEANHAMNCALNDWQANHWLDSHNNWHER WRGSICLAVEAPELAAQEIERWAGHPYFAQCLIKAEPRPSWGDPKYDPLWAAATKHDIVVSCHLSRGEYD ELPTPPVGLPSYNHDFMDAIYERRKSWMDIKRKPSEYVKDHIKFTTQPLDYPEDKTELSRAFEWMECEKI LLFSSDYPHWTFDDPRWLVKHLPEHAREAVMFRNGIETYKLPDTVPVLEGQVRVF
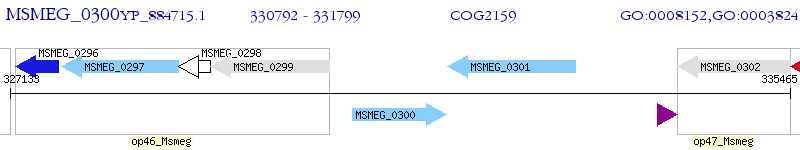
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0300 | - | - | 100% (335) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4808 | - | e-161 | 73.08% (364) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4807 | - | 1e-27 | 27.75% (346) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2489 | - | 1e-175 | 75.79% (380) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4725 | - | 1e-158 | 68.97% (377) | metal-dependent hydrolase |
| M. avium 104 | MAV_0929 | - | 1e-157 | 70.21% (376) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_0524 | - | 1e-171 | 74.21% (380) | amidohydrolase family protein |
| M. ulcerans Agy99 | MUL_3849 | - | 5e-06 | 27.61% (134) | metal-dependent hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4165 | - | 1e-175 | 76.05% (380) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4725|M.marinum_M MTLTHMHERVPAAERIAVRCVDSDIHPVP-KRGEISPYIPEPWRSKFFLD
MAV_0929|M.avium_104 MTLTHMQERVPAAERIAVRCVDSDVHPVP-KRGEITPYIPERWR-KFFFE
Mflv_2489|M.gilvum_PYR-GCK MTVT-ANPRVPAAERIAVRCVDSDVHPAP-RRGELTPYIPEPWRSKYFLT
Mvan_4165|M.vanbaalenii_PYR-1 MTVT-ANPRVAATERIAVRCVDSDVHPTP-RRGELLPYIPEPWRSKYFLT
TH_0524|M.thermoresistible__bu MTVT-TNPRVPATERTAIRCVDSDVHPTP-RRGELLPYIPEPWRSKYFLT
MSMEG_0300|M.smegmatis_MC2_155 MTIH-TSQRTPAAERIAVRCVDSDVHPVP-RRGVLGEYIPEPWRSSYFGT
MUL_3849|M.ulcerans_Agy99 MPSRELPFPVFDADNHMYEPQEALTKFLPDKRKHVIDYVQVRGRTKIVVR
*. . ::. . :: : * :* : *: * . .
MMAR_4725|M.marinum_M HKVGELIYYDAPDYAHSFAMRVDTFPADG------EFPGS--DPDMAFR-
MAV_0929|M.avium_104 HRVGELIYYDAPDYAHAFAMRTDTFPPDG------EFPGS--DPDMAFR-
Mflv_2489|M.gilvum_PYR-GCK RKIGEQIYYDAPDYAHSFAMRVDAFPSDG------EFACS--DPDLAFK-
Mvan_4165|M.vanbaalenii_PYR-1 RKVGEQIYYDAPDYAHSYAMRVDTFPPDG------EFACS--DPDLAFK-
TH_0524|M.thermoresistible__bu RKVGEQIYYDAPDYAHSYAMRVDAFPPDG------EFACS--DPDLAFR-
MSMEG_0300|M.smegmatis_MC2_155 HDVGELIYYDAPDYAHAYAMRVDTFPSDG------EFAGS--DPDLAFR-
MUL_3849|M.ulcerans_Agy99 GQISDYIPNPTFEVVARPGAQEEHFRNGSGGKSYREVMGEPMKAIPAFRE
:.: * : : . . : : * .. *. . .. **:
MMAR_4725|M.marinum_M -----QLIMEAGSDIAILEP------GGRTPRLPEAHQAYSTALNRWQAN
MAV_0929|M.avium_104 -----QLIMEAGSDIAILEP------AGRTPRIGEAHQAYCSALNDWQAN
Mflv_2489|M.gilvum_PYR-GCK -----QLIMEAGADIAILEP------AAYPARIPEAQHAMSCALNDWQAN
Mvan_4165|M.vanbaalenii_PYR-1 -----QLIMEAGADIAVLEP------AAYPARIPEAQHAMSVALNDWQAN
TH_0524|M.thermoresistible__bu -----QLIMEAGADIAILEP------AAYPARTPETQHAMAVALNDWQAN
MSMEG_0300|M.smegmatis_MC2_155 -----QLIMEAGADIAILEP------AAYSAHIPEANHAMNCALNDWQAN
MUL_3849|M.ulcerans_Agy99 PGPRLEVMDELGLDYALMFPTLASLVEERMKDDPEMTHDVIHALNQWMYE
::: * * * *:: * * : *** * :
MMAR_4725|M.marinum_M HWLDSHNNWHQRWRGSICAAVEDPEGAAREIEEWAGHPYMAQVLIKAEPR
MAV_0929|M.avium_104 HWLDSRNNWHERWRGSICAAIEDPEGAVEQIEKWAGHPYMAQILIKAEPR
Mflv_2489|M.gilvum_PYR-GCK HWLDSHNNWHERWRGSICLAIEEPEHSVAEIERWVGHPYMAQILIKAEPR
Mvan_4165|M.vanbaalenii_PYR-1 HWLDSHNNWHERWRGSICLAIEEPEKSVEEIERWAGHPYMAQILIKAEPR
TH_0524|M.thermoresistible__bu HWLDSHNNWHQRWRGSICLAIEAPEDSVREIERWAGHPYMAQILIKAEPR
MSMEG_0300|M.smegmatis_MC2_155 HWLDSHNNWHERWRGSICLAVEAPELAAQEIERWAGHPYFAQCLIKAEPR
MUL_3849|M.ulcerans_Agy99 TWS---FNYKDRIFATPVITLPIVDRALEELE-WCLQRGARTVLVRPAPV
* *:::* .: :: : : ::* * : *::. *
MMAR_4725|M.marinum_M P------CWGHPMYNPIWAAATKHDITVSCHLSRSNFEMLP---TPPVGY
MAV_0929|M.avium_104 P------SWGHPKYDPIWAAATKHDIVVSCHLSRSNYEMLP---TPPVGF
Mflv_2489|M.gilvum_PYR-GCK P------SWGNPKYDPIWAAAAKHDIPVSCHLSRSHYDELP---MPPVGL
Mvan_4165|M.vanbaalenii_PYR-1 P------SWGNPKYDPIWAAATKHDITVSCHLSRSHYDELP---MPPVGL
TH_0524|M.thermoresistible__bu P------SWGDPKYDPIWAAAAKHDITVSCHLSRSHHEELP---MPPVGM
MSMEG_0300|M.smegmatis_MC2_155 P------SWGDPKYDPLWAAATKHDIVVSCHLSRGEYDELP---TPPVGL
MUL_3849|M.ulcerans_Agy99 PGYRGSRSFGFKEFDPFWQACIKAGIPVSMHASDSGYSEFLNVWEPGDEF
* .:* ::*:* *. * .* ** * * . .. : *
MMAR_4725|M.marinum_M PSYN---HDFMVTYSLLAANQVMSLIFDGVFDRFPTLRIVLVEHAFTWIL
MAV_0929|M.avium_104 PSYN---HDFMVTYSLLAANQVMSLIFDGVFDRFPTLRIVFVEHAFTWIL
Mflv_2489|M.gilvum_PYR-GCK PSYN---HDFMVTYSLLAANQVMSLIFDGTFDRHPNLRIVFVEHAFTWIL
Mvan_4165|M.vanbaalenii_PYR-1 PSYN---HDFMVTYSLLAANQVMSLIFDGTFDRHPNLRIVFVEHAFTWIL
TH_0524|M.thermoresistible__bu PSYN---HDFMVTYSLLAVNQVMSLIFDGVFDRFPNLRVVFVEHAFTWIL
MSMEG_0300|M.smegmatis_MC2_155 PSYN---HDF----------------------------------------
MUL_3849|M.ulcerans_Agy99 LPFKPTAFRMLAMSKRPIEDAMGALVCHGALSRNPELRILSIKNGADWVP
.:: . :
MMAR_4725|M.marinum_M PLMWRMDALYDARKSRMDIKRKPSEYVKEHIKFTTQPLDYPEDKTELTRA
MAV_0929|M.avium_104 PLMWRMDAIYEARKSRVQIKRKPSEYVKDHIKFTTQPLDYPEDKTELTRA
Mflv_2489|M.gilvum_PYR-GCK PLMWRMDALYEARKSWVEIKRKPSEYVKDHIKFTTQPLDYPEDKTELSRA
Mvan_4165|M.vanbaalenii_PYR-1 PLMWRMDALYEARKSWVDIKRRPSEYVKDHIKFTTQPLDYPEDKTELSRA
TH_0524|M.thermoresistible__bu PLMWRMDAIYEARKSWLEIKRKPSEYVKDHIKFTTQPLDYPEDKTELTRA
MSMEG_0300|M.smegmatis_MC2_155 -----MDAIYERRKSWMDIKRKPSEYVKDHIKFTTQPLDYPEDKTELSRA
MUL_3849|M.ulcerans_Agy99 TLFKGLKGVY---------KKMPNAFMEDPIEAFKRCVYISPFWEDRFVE
:..:* *: *. :::: *: .: : . :
MMAR_4725|M.marinum_M IEWMEGDKILLYSSDYPHWT-FDDPRWLVKHLPKAARDAVMYKNGIATYH
MAV_0929|M.avium_104 LEWMECDKILLYSSDYPHWT-FDDPRWLVKHLPKAARDAVMYKNGIATYH
Mflv_2489|M.gilvum_PYR-GCK FEWMECEKILLFSSDYPHWT-FDDPRWLVKHLPEHAREAVMFKNGIETYK
Mvan_4165|M.vanbaalenii_PYR-1 FEWMECEKILLFSSDYPHWT-FDDPRWLVKHLPEHAREAVMFRNGIETYK
TH_0524|M.thermoresistible__bu LEWMECEKILLFSSDYPHWT-FDDPRWLVKHLPEHAREAVMFRNGIETYK
MSMEG_0300|M.smegmatis_MC2_155 FEWMECEKILLFSSDYPHWT-FDDPRWLVKHLPEHAREAVMFRNGIETYK
MUL_3849|M.ulcerans_Agy99 IVKMVGTDRVVFGSDWPHPEGLKDPISFVDELAGFPPEDVAKIMGGNMMD
: * . :::.**:** :.** :*..*. . : * * .
MMAR_4725|M.marinum_M LPDTVPVLEGQTRVF
MAV_0929|M.avium_104 LPETVPVLEGQVRVL
Mflv_2489|M.gilvum_PYR-GCK LPDTVPALEGQVRVF
Mvan_4165|M.vanbaalenii_PYR-1 LPDTVPALEGQVRVF
TH_0524|M.thermoresistible__bu LPETVPALEGQTRVF
MSMEG_0300|M.smegmatis_MC2_155 LPDTVPVLEGQVRVF
MUL_3849|M.ulcerans_Agy99 VMKIGTAAPKPVNA-
: . .. ...