For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSRELPFPVFDADNHMYEPQEALTKFLPDKRKHVIDYVQVRGRTKIVVRGQISDYIPNPTFEVVARPGA QEEHFRNGSGGKSYREVMGEPMKAIPAFREPGPRLEVMDELGLDYALMFPTLASLVEERMKDDPEMTHDV IHALNQWMYETWSFNYKDRIFATPVITLPIVDRALEELEWCLQRGARTVLVRPAPVPGYRGSRSFGFKEF DPFWQACIKAGIPVSMHASDSGYSEFLNVWEPGDEFLPFKPTAFRMLAMSKRPIEDAMGALVCHGALSRN PELRILSIKNGADWVPTLFKGLKGVYKKMPNAFMEDPIEAFKRCVYISPFWEDRFVEIVKMVGTDRVVFG SDWPHPEGLKDPISFVDELAGFPPEDVAKIMGGNMMDVMKIGTAAPKPVNA
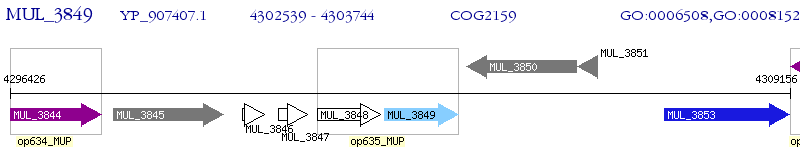
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3849 | - | - | 100% (401) | metal-dependent hydrolase |
| M. ulcerans Agy99 | MUL_0308 | - | e-136 | 57.87% (394) | hypothetical protein MUL_0308 |
| M. ulcerans Agy99 | MUL_3878 | - | 4e-79 | 38.44% (398) | metal-dependent hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3792 | - | 1e-178 | 72.52% (393) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0299 | - | 1e-07 | 24.32% (292) | putative amidohydrolase (aminocarboxymuconate-semialdehyde |
| M. marinum M | MMAR_3988 | - | 0.0 | 97.51% (401) | metal-dependent hydrolase |
| M. avium 104 | MAV_1625 | - | 0.0 | 88.56% (402) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_2994 | - | 0.0 | 75.06% (393) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_0536 | - | 0.0 | 75.57% (393) | amidohydrolase family protein |
| M. vanbaalenii PYR-1 | Mvan_2602 | - | 0.0 | 75.32% (393) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3849|M.ulcerans_Agy99 MPSRELPFPVFDADNHMYEPQEALTKFLPDKRKHVIDYVQVRGRTKIVVR
MMAR_3988|M.marinum_M MPSRELPFPVFDADNHMYEPQEALTKFLPDKRRHVIDYVQVRGRTKIVVR
MAV_1625|M.avium_104 MPSRELSFPVFDADNHMYEPQEALTKFLPDHRKHVIDYVQIRGRTKIVVR
Mflv_3792|M.gilvum_PYR-GCK MPSRSLDYPVFDADNHFYEPKEALTQFLPDHRKGVIDYIDVHGRTKIVVR
TH_0536|M.thermoresistible__bu MPSRELPYPVFDADNHFYEPQEALTKFLPEHRKGVIDYIDVRGRTKIVVR
MSMEG_2994|M.smegmatis_MC2_155 MPSRILDFPVFDADNHFYEPKEALTKFLPDHRKGVIDYVDIRGRTKIVVR
Mvan_2602|M.vanbaalenii_PYR-1 MPSRILDYPVFDADNHFYEPKEALTKFLPDKRKGVIDYIDVHGRTKIMVR
MAB_0299|M.abscessus_ATCC_1997 -----MTSVRIDIHAHLWS-------------------------------
: :* . *::.
MUL_3849|M.ulcerans_Agy99 GQISDYIPNPTFEVVARPGAQEEHFRNGSGGKSYREVMGEPMKAIPAFRE
MMAR_3988|M.marinum_M GQISDYIPNPTFEVVARPGAQEEYFRNGSRGKSYREVMGEPMKAIPAFRE
MAV_1625|M.avium_104 GHISEYIPNPTFEVVARPGAQEEYFRNGSGGKSYREILGEPMKAIPAFRE
Mflv_3792|M.gilvum_PYR-GCK NHISDYIPNPTFEVVARPGAQEEYFKHGSGGKSFREVMGKPMKAIPAFRN
TH_0536|M.thermoresistible__bu NQISDYIPNPTFEVVARPGAQEEYFKYGSGGKSFREVMGKPMKAIPAFRD
MSMEG_2994|M.smegmatis_MC2_155 NHISDYIPNPTFEVVARPGAQEEYFRQGSGGKSYREVLGKPMKSIPAFRN
Mvan_2602|M.vanbaalenii_PYR-1 NHISDYIPNPTFEVVARPGAQEEYFRHGSGGKSFREVMGKPMKAIPAFRN
MAB_0299|M.abscessus_ATCC_1997 ---DEYLS-----LLDGYGRPATDVHRGLGAGATRDDLDG----------
.:*:. :: * .: * . : *: :.
MUL_3849|M.ulcerans_Agy99 PGPRLEVMDELGLDYALMFPTLASLVEERMKDDPEMTHDVIHALNQWMYE
MMAR_3988|M.marinum_M PGPRLEVMDELGLDYALMFPTLASLVEERMKDDPEMTHDVIHALNQWMYE
MAV_1625|M.avium_104 PGARLEVMDELGIDYALMFPTLASLVEERMKDDPEMTHDVIHALNQWMYE
Mflv_3792|M.gilvum_PYR-GCK PEARLEVLDGLGLDYTIMFPTLASLVEERMKDDPDLILDIVHALNQWMYE
TH_0536|M.thermoresistible__bu PEARLEVLDGLGLDYTIMFPTLASLVEERMKDDPDLILDVIHALNQWMYE
MSMEG_2994|M.smegmatis_MC2_155 PEARLEVMDGLGLDYTLMFPTLASLVEERLKDDPELIHDIIHALNEWMYE
Mvan_2602|M.vanbaalenii_PYR-1 PEARLEVMDGLGLDYTLMFPTLASLVEERLKDDPELIHDIIHALNEWMYE
MAB_0299|M.abscessus_ATCC_1997 ---RFELMDEAGVDLQILTATPAS---PHFEDQAAAVASARFIN--DEYA
*:*::* *:* :: .* ** :::*:. . . *
MUL_3849|M.ulcerans_Agy99 TWSFNYKDRIFATPVITLPIVDRALEELEWCLQRGARTVLVRPAPVPGYR
MMAR_3988|M.marinum_M TWSFNYKDRIFATPVITLPIVDRALEELEWCLQRGARTVLVRPAPVPGYR
MAV_1625|M.avium_104 QWSFNYKDRIFATPVITLPIVDRALEELEWCLERGARTVLVRPAPVPGYR
Mflv_3792|M.gilvum_PYR-GCK TWQFDYQGRILSTPVINLSVVDRALEELQWCLERGARTVLVRPAPVPGYR
TH_0536|M.thermoresistible__bu TWQFDYEGRILSTPVINLSVVDRALEELEWCLERGAKTILVRPAPVPGYR
MSMEG_2994|M.smegmatis_MC2_155 TWQFDYEGRIFSTPVITLPIVDRALEELEWCLERGAKTVLVRPAPVPGFR
Mvan_2602|M.vanbaalenii_PYR-1 TWQFDYEGRIFSTPVITLPIVDRALEELEWCLERGAKTVLVRPAPVPGFR
MAB_0299|M.abscessus_ATCC_1997 KLSSEYPGRFGALASLPLPHVPAALEELARAIDELGMYGASITTSVLG--
. :* .*: : . : *. * ***** .::. . .:.* *
MUL_3849|M.ulcerans_Agy99 GSRSFGFKEFDPFWQACIKAGIPVSMHASDSGYSEFLNVWEPGDEFLPFK
MMAR_3988|M.marinum_M GSRSFGFEEFDPFWQACIKAGIPVSMHASDSGYSEFLNVWEPGDEFLPFK
MAV_1625|M.avium_104 GSRSFGFEEFDPFWQACVRAEIPVSMHASDSGYSELLNIWEPGDEFLPFK
Mflv_3792|M.gilvum_PYR-GCK GSRSLGLPEFDPFWDACVQAGIPVCMHASDSGYAQYLNDWEPADEFLPFR
TH_0536|M.thermoresistible__bu GTRSLGLPEFDPFWDACVKAGIPVCMHASDSGYAQYLNDWEPADEFLPFR
MSMEG_2994|M.smegmatis_MC2_155 GTRSFGTEEFDPFWQACVKAGIPVAMHASDSGYAQYLNDWEPADEFLPFK
Mvan_2602|M.vanbaalenii_PYR-1 GTRSFGTEEFDPFWDACVKAGIPVAMHASDSGYAQYLNDWEPADEFLPFR
MAB_0299|M.abscessus_ATCC_1997 --RSIADPIFDPLYAELNRRRAVLFVHPAGCGAESSL----ITDHSLTWS
**:. ***:: : : :*.:..* . * *. *.:
MUL_3849|M.ulcerans_Agy99 PTAFRMLAMSKRPIED--AMGALVCHGALSRNPELRILSIKNGADWVPTL
MMAR_3988|M.marinum_M PTAFRMLAMGKRPIED--AMGALVCHGALSRNPELRILSIENGADWVPTL
MAV_1625|M.avium_104 PTAFRSLAMGHRPVED--AFGALICHGALSRNPDLRILSIENGADWVPHL
Mflv_3792|M.gilvum_PYR-GCK PTAFRMVAMGKRPIED--TMAALVCHGALTRNPDLRILSIENGASWVPYL
TH_0536|M.thermoresistible__bu PTAFRMVAMGKRPIED--TMAALVCHGALSRNPDLRILSVENGASWVPYL
MSMEG_2994|M.smegmatis_MC2_155 PTAFRMVAMGKRPIED--TMTALVCHGTFTRNPDLRILSIENGASWVPHL
Mvan_2602|M.vanbaalenii_PYR-1 PTAFRMVAMGKRPIED--TMAALVCHGAFTRNPDLRILSIENGASWVPYL
MAB_0299|M.abscessus_ATCC_1997 --------IG-APIEDTVAIMHLIVAGVPSRYPDMKIVTCHLGGALPMVL
:. *:** :: *: *. :* *:::*:: . *. *
MUL_3849|M.ulcerans_Agy99 FKGLKGVYKKMPNAFMEDPIEAFKRCVYISPFWEDR--FVEIVKMVGTDR
MMAR_3988|M.marinum_M FKGLKGVYKKMPNAFMEDPIEAFKRCVYISPFWEDR--FAEIVKMVGTDR
MAV_1625|M.avium_104 FKGLKGVYKKMPQAFAEDPIETFKRCVYVSPFWEDR--FAEIVNMVGTDR
Mflv_3792|M.gilvum_PYR-GCK FYQFEDVYKKMPQEFPENPIDAFRRGVYVAPFWEDD--FGKMADLLGIDR
TH_0536|M.thermoresistible__bu FYQFKDVYKKMPQEFAEDPIEAFKRCVYVAPFWEDN--FKEIADLIGIDR
MSMEG_2994|M.smegmatis_MC2_155 FHQFEDVYAKMPQDFPEEPIQAFKRCVYVAPFWEDN--FKQMADLIGIDR
Mvan_2602|M.vanbaalenii_PYR-1 FYQFKDVYSKMPQEFPEDPIEAFRRCVYVAPFWEDD--FKKMADLCGIDR
MAB_0299|M.abscessus_ATCC_1997 ERAHRQVTEWEATQCPEAPRDAARRLWYDTVGHDHTPALRAAVASLGVDR
. * . * * :: :* * : :. : . * **
MUL_3849|M.ulcerans_Agy99 VVFGSDWPHPEGLKDPISFVDELAG--FPPEDVAKIMGGNMMDVMKIGTA
MMAR_3988|M.marinum_M VVFGSDWPHPEGLKDPISFVDELAG--FPPEDVAKIMGGNMMEVMKIGTP
MAV_1625|M.avium_104 VVFGSDWPHPEGLKDPISFVDELAD--FSDEDKAKIMGGNLMKLMKVSAP
Mflv_3792|M.gilvum_PYR-GCK VIFGSDWPHPEGLADPITLVDQLEENGLDEDGIRKVMGGNLVDLFKVPNK
TH_0536|M.thermoresistible__bu VIFGSDWPHPEGLADPIRLVDELEEHGLDDEGIRKVMGANLIELFGVENK
MSMEG_2994|M.smegmatis_MC2_155 VIFGSDWPHPEGLADPINLVSDLQAHGLDDEGVRKVMGRNLVDLFQVENK
Mvan_2602|M.vanbaalenii_PYR-1 VIFGSDWPHPEGLADPINLVADLEAHGLDQEGVRKVMGKNMVDLFRVENK
MAB_0299|M.abscessus_ATCC_1997 LIFGSDFPYQAGPRYVSSATYIEEG--LSAEDASAILDHNGAELLGLTGV
::****:*: * . : :. ::. * .:: :
MUL_3849|M.ulcerans_Agy99 AP-KPVNA------
MMAR_3988|M.marinum_M AP-QPVNA------
MAV_1625|M.avium_104 AAKKPVSA------
Mflv_3792|M.gilvum_PYR-GCK VVHKPDAPALVIA-
TH_0536|M.thermoresistible__bu KVHNPNVPALEIPA
MSMEG_2994|M.smegmatis_MC2_155 VVYKPDVPALVIA-
Mvan_2602|M.vanbaalenii_PYR-1 VVYKPDVPALVIA-
MAB_0299|M.abscessus_ATCC_1997 --------------