For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHIFFGSNKDLRQNFLREPFASRGFPDYEMNWYGAPGGEYAKNTKGPDRQYPGSDPDVAAQHLFTERGVD IAILHPMTRGIMPDRHLGTALANAHNEMMVTRWLDHERYGERFRGTIRVNPDDIAGALREIERFKDHPRV VQIGVPLQSRELYGKPQYWPLWEAASDAGLPVAVHIEVGAGVQFAPTPSGKTRTYEQYLGFMALNYLYHL MNMIAEGVFERMPALKFVWADGAADLLTPFIWRMDCFGRPHLEQTPWAPKMPSDYLPGHVYFVQGALDGP GDVEFAGEWFGFTGKENMVMFGSSYPHWQLNEPEVPTAFTPEQRDKLLWRNAAELYGLQSALTSTAGAAR
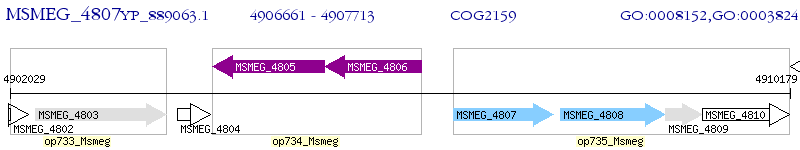
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4807 | - | - | 100% (350) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4808 | - | 3e-40 | 30.00% (350) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_0300 | - | 1e-27 | 27.75% (346) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2490 | - | 0.0 | 80.86% (350) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4724 | - | 1e-167 | 76.35% (351) | metal-dependent hydrolase |
| M. avium 104 | MAV_0930 | - | 1e-168 | 77.23% (347) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_4618 | - | 0.0 | 84.00% (350) | PUTATIVE amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_4959 | - | 4e-05 | 23.85% (327) | hypothetical protein MUL_4959 |
| M. vanbaalenii PYR-1 | Mvan_4164 | - | 0.0 | 81.82% (352) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4724|M.marinum_M MVIEHADGTRM--------PVIDASVHIFFPSNK--------DLR-SFLR
MAV_0930|M.avium_104 -MIEHADGTRT--------PVIDASVHIFFPSNK--------DLR-SFLR
Mflv_2490|M.gilvum_PYR-GCK -MKEFSDGSSG----VARVPVIDASVHIFCQSNK--------DLRKNFLR
Mvan_4164|M.vanbaalenii_PYR-1 -MKVLSDGSAGALEAQVKVPVIDASVHIFCQSNK--------DLRRNFLQ
MSMEG_4807|M.smegmatis_MC2_155 -------------------------MHIFFGSNK--------DLRQNFLR
TH_4618|M.thermoresistible__bu -VIELSDGTRT--------PVIDASVHIFCKSNK--------DLRGTFLR
MUL_4959|M.ulcerans_Agy99 -MKTELGFTFFDCGNHYYEAVDAFTRHIEREYKKRAIQWAQLDGRARLII
** :* * * ::
MMAR_4724|M.marinum_M EPFKSRGFPDYEMDWYGAPGSE----YAPNTEGPDRQYPGSDPKFVADEL
MAV_0930|M.avium_104 EPFKSRGFPDYEMDWYGAPGSE----YAPNTDGPDGQYPGSDPEFVAQEL
Mflv_2490|M.gilvum_PYR-GCK EPFRSRGFPDYEMDWYGAPGGE----YAPRTEGPDGQYPGSDPELVAKHL
Mvan_4164|M.vanbaalenii_PYR-1 EPFRSRGFPDYEMDWYGAPGGE----YAERTTGPDDQYPGSDPEVVAKHL
MSMEG_4807|M.smegmatis_MC2_155 EPFASRGFPDYEMNWYGAPGGE----YAKNTKGPDRQYPGSDPDVAAQHL
TH_4618|M.thermoresistible__bu EPFKSRGFPDYEMDWYGAPGGE----YAKGTEGPDRQYPGSDPDVVAQHL
MUL_4959|M.ulcerans_Agy99 GGRVNRFIPNPTFDPVGKPGALDEYFRGRNPRGADLNALFGDLEPIRPEY
.* :*: :: * **. . . *.* : .* . .
MMAR_4724|M.marinum_M FS---------RRGVDVAVLHP-----MGRGIMPDRHLGSALHAAHNEMM
MAV_0930|M.avium_104 FS---------KRGVDVAVLHP-----MGRGIMPDRHLGSALHAAHNEMM
Mflv_2490|M.gilvum_PYR-GCK FT---------DRGVDLAILHP-----MTRGIMPDRHLGTAIASAHNEMM
Mvan_4164|M.vanbaalenii_PYR-1 FT---------DRGVDIAVLHP-----MTRGIMPDRHLGTALAAAHNEMM
MSMEG_4807|M.smegmatis_MC2_155 FT---------ERGVDIAILHP-----MTRGIMPDRHLGTALANAHNEMM
TH_4618|M.thermoresistible__bu FE---------QRGVDIAVLHP-----MTRGIMPDRHLGTALAAAHNEML
MUL_4959|M.ulcerans_Agy99 RDRDARLAAMDEQGMQGCIMLPTLGVGMEQALLPDLEATAATFRAFNRWM
:*:: .:: * * :.::** . :* *.*. :
MMAR_4724|M.marinum_M VSRWLEHDEFGDRFRGTIRVNPDDIAGAVREVAKWRAHPR--VVQIGVPL
MAV_0930|M.avium_104 VSRWLDSPEFGEKFRGTIRVNPDDIAGALRELERWRSHPR--VVQLGIPL
Mflv_2490|M.gilvum_PYR-GCK VTRWLDHPEFGERFRGTLRVNPDDIAGALREIDKYRDHPR--IVQLGIPL
Mvan_4164|M.vanbaalenii_PYR-1 VTRWLDHPQLGEKFRGTLRVNPDDIAGALREIDKYKSHPR--IVQLGIPL
MSMEG_4807|M.smegmatis_MC2_155 VTRWLDHERYGERFRGTIRVNPDDIAGALREIERFKDHPR--VVQIGVPL
TH_4618|M.thermoresistible__bu VTRWLDHPEFGDRFRGTIRVNPDDIAGSLREIEKYKDHPR--VVQLGIPL
MUL_4959|M.ulcerans_Agy99 QEDWG--FAYQERIFAAPYITLCIPDNAVRELEWALRHDARFIVMVPGPV
* ::: .: :. .::**: * :* : *:
MMAR_4724|M.marinum_M QSRE---LYGKPQFWPLWEAAVEAGLPVAVHIESGEGIGFPPTP--SGHT
MAV_0930|M.avium_104 QSRE---LYGKPQFWPLWEAAADANLPVAVHIETGEGIGFPPTP--SGHT
Mflv_2490|M.gilvum_PYR-GCK QSRE---VYGKPQFWPLWEAAVDAGWPVAVHFEVGSGVALPPTP--NGLT
Mvan_4164|M.vanbaalenii_PYR-1 QSRE---LYGKPQYWPLWEAAVDAGWPVAVHFEVGSGVQLPPTP--SGLT
MSMEG_4807|M.smegmatis_MC2_155 QSRE---LYGKPQYWPLWEAASDAGLPVAVHIEVGAGVQFAPTP--SGKT
TH_4618|M.thermoresistible__bu QSRE---LYGKPQFWPLWEAAVAANLPVAVHIEVGAGIAASPTP--SGNT
MUL_4959|M.ulcerans_Agy99 TTEVGMRAPGDPLFDPFWQLANDSGITVCYHSGETYYSKFMPAWGEADYM
:. *.* : *:*: * :. .*. * *: .
MMAR_4724|M.marinum_M RTYEQYVSFMALN----YLYHLMNMIAEGVFERFAELKFIWADGAADFVT
MAV_0930|M.avium_104 QTYEQYVSFMALN----YLYHLMNMIAEGVFERFPALKFVWADGAADFLT
Mflv_2490|M.gilvum_PYR-GCK RTYEQYVGFTALN----FLYHLMNMIAEGVFERMPDLKFVWADGAADLLT
Mvan_4164|M.vanbaalenii_PYR-1 RTYEQFVGFTALN----FLYHLMNMIAEGVFERMPALKFVWADGAADMLT
MSMEG_4807|M.smegmatis_MC2_155 RTYEQYLGFMALN----YLYHLMNMIAEGVFERMPALKFVWADGAADLLT
TH_4618|M.thermoresistible__bu RTYEQYVGFMALN----YLYHLMNMIAEGVFERMPTLKFVWADGAADLLT
MUL_4959|M.ulcerans_Agy99 MSFQALLGFRSLFSGDPLQDTFANHLHKGLFHRFPNLRMACIESGSAWVF
::: :.* :* : * : :*:*.*:. *:: :..: :
MMAR_4724|M.marinum_M PFIWRMDTFGRPHLEQTPWAPR-IPSDYLPGHVYFIQGGLDGPGDVEFAD
MAV_0930|M.avium_104 PFIWRMDTFGRPHLEQTPWAPR-IPSDYIPGHVYFVQGSLDGPGDADFAA
Mflv_2490|M.gilvum_PYR-GCK PFMWRMDCFGRPHLEQTPWAPR-MPSDYLPGHVFFVQGALDGPGDVDFAG
Mvan_4164|M.vanbaalenii_PYR-1 PFIWRMDCFGRPHLEQTPWAPR-IPSDYLPDHVYFVQGALDGPGDVDFAG
MSMEG_4807|M.smegmatis_MC2_155 PFIWRMDCFGRPHLEQTPWAPK-MPSDYLPGHVYFVQGALDGPGDVEFAG
TH_4618|M.thermoresistible__bu PFIWRMDCFGRPHLEQTPWAPR-MPSDYLPGHVYFVQGFLDGPGDVDFAG
MUL_4959|M.ulcerans_Agy99 HLFEKLAKS----YGQIPFIYQEDPRETFKRHVWVS------PFYEDELA
:: :: * *: : * : : **:. * :
MMAR_4724|M.marinum_M EWLSFTGKEDMVMFGSSYPHWQCYDLRDVRNLPTALTADQREKLCWRNAA
MAV_0930|M.avium_104 EWFGFTGKEDMVMFGSSYPHWQC---ADVHKLPPALSAEQREKLCWRTAA
Mflv_2490|M.gilvum_PYR-GCK EWAGFTGKDDMVMYGSSYPHWQL----NELTVPGSYDPEQRDKLCWRNAA
Mvan_4164|M.vanbaalenii_PYR-1 EWAGFTGKDDMVMYGSSYPHWQL----NELSVPTAYTPDQRDKLCWRNAA
MSMEG_4807|M.smegmatis_MC2_155 EWFGFTGKENMVMFGSSYPHWQL----NEPEVPTAFTPEQRDKLLWRNAA
TH_4618|M.thermoresistible__bu EWFGFTGKENMVMFGSSYPHWQL----NELTIPGTFSAEQRDKLCWRNAA
MUL_4959|M.ulcerans_Agy99 DLLKLLG-ADRIIMGSDYPHVEG------LAEPASYIKDLQNFDYAPDQC
: : * : :: **.*** : * : : :: .
MMAR_4724|M.marinum_M ELYGIDI------SAELTSG-
MAV_0930|M.avium_104 DLYGIDI------SVDLAAS-
Mflv_2490|M.gilvum_PYR-GCK QLYGIQS---PAISVPAAAQ-
Mvan_4164|M.vanbaalenii_PYR-1 ALYGIESPVIPAVSTTAAAQ-
MSMEG_4807|M.smegmatis_MC2_155 ELYGLQS---ALTSTAGAAR-
TH_4618|M.thermoresistible__bu ELYNIES---SAIPSAALAK-
MUL_4959|M.ulcerans_Agy99 RAVMRDN----GLELALRRPV
: