For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSNQRPLEGRRALVTGGTKGAGAAVVRTLRASGAHVTAVARRPGGDADEFVAADVMSEAGIAEIATHVRA TGGVHILVHVAGGSSSPAGGFAALSEQDWIDELNLNLLSAVRLDRAVAPMMIEAGGGVIVHISSIQSRMP LYDGTLGYAAAKAALRTYSKGLANELAPKGIRVNTVSPGFIRTTAAEALIERIAEAGNGTREQALDSLMA SLGGIPLGRPAEPDEVAGVVGFLVSDAAASVVGADIVVDGGTVPAI
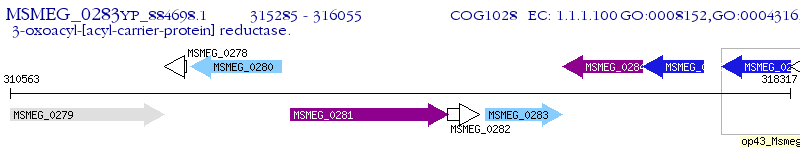
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0283 | - | - | 100% (256) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3607 | - | 1e-31 | 39.00% (259) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5204 | - | 7e-30 | 36.84% (266) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 4e-29 | 37.88% (264) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0943 | - | 1e-42 | 39.13% (253) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 4e-29 | 37.88% (264) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-22 | 33.88% (245) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0607 | - | 3e-30 | 38.46% (247) | short chain dehydrogenase |
| M. marinum M | MMAR_2981 | fabG3_1 | 2e-30 | 37.88% (264) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_2509 | - | 9e-28 | 36.74% (264) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_3911 | - | 9e-31 | 33.72% (258) | PUTATIVE putative short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 1e-23 | 33.46% (257) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_5918 | - | 9e-41 | 37.94% (253) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
MMAR_2981|M.marinum_M --------------MAGRLTGKVALVSGGARGMGASHVRALVAEGAHVVL
MAV_2509|M.avium_104 --------------MAERLAGKVALVSGGARGMGASHVRSLVAEGAKVVF
MUL_1037|M.ulcerans_Agy99 ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MSMEG_0283|M.smegmatis_MC2_155 ------------MSNQRPLEGRRALVTGGTKGAGAAVVRTLRASGAHVTA
MAB_0607|M.abscessus_ATCC_1997 ----------MRTPYDLGLEGRVVLVTGGVRGVGAGISQVFADQGATVVT
Mflv_0943|M.gilvum_PYR-GCK -MSSTIVAPSVDATPIQDLAGKTAIITGGSKGIGAATVARFVRGGARVVT
Mvan_5918|M.vanbaalenii_PYR-1 -MNSAIVAPEVPTAPIHDLTGKNAVITGGSKGIGAATVARFVAGGARVVT
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
TH_3911|M.thermoresistible__bu -----------------MTKPRNALVTGGTDGIGKAIALRLRADGMRVAV
: :::** * * . : * *.
Mb2025|M.bovis_AF2122/97 GDILDE--EGKAVAAELADAARYVHLDVTQPAQWTAAVDTAVTAFGGLHV
Rv2002|M.tuberculosis_H37Rv GDILDE--EGKAVAAELADAARYVHLDVTQPAQWTAAVDTAVTAFGGLHV
MMAR_2981|M.marinum_M GDILDD--EGRAVAAELGDAARYVHLDVTQPEQWTAAVDTAVNEFGGLHV
MAV_2509|M.avium_104 GDILDD--EGKAVAAEVGEATRYLHLDVTKPEDWDAAVATALAEFGRIDV
MUL_1037|M.ulcerans_Agy99 GDILDE--EGKALAEEIGDAARYVHLDVTQPDQWEAAVATAVDEFGKLDV
MSMEG_0283|M.smegmatis_MC2_155 --------VARRPGG---DADEFVAADVMSEAGIA-EIATHVRATGGVHI
MAB_0607|M.abscessus_ATCC_1997 --------CARRPGD---APYEFHSCDVRDEESVDTLIGAIVAAHGRLDA
Mflv_0943|M.gilvum_PYR-GCK --------SGRSTPGPLPDGVEYVVADVATVDGVEHLARRAQQILGDIDI
Mvan_5918|M.vanbaalenii_PYR-1 --------SGRSQPDSLPTGVEYVVADVATLDGVDLLARRAREILGDIDI
MLBr_01807|M.leprae_Br4923 THR----------ASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPVEV
TH_3911|M.thermoresistible__bu TGRNAERGAQLVDEVGCPEEFVFIRSDATDPHDAERVAAEVRNALGGIGV
*. * :
Mb2025|M.bovis_AF2122/97 LVNNAGI--LN-IGTIEDYALTEWQRILDVN-LTGVFLGIRAVVKPMKEA
Rv2002|M.tuberculosis_H37Rv LVNNAGI--LN-IGTIEDYALTEWQRILDVN-LTGVFLGIRAVVKPMKEA
MMAR_2981|M.marinum_M LVNNAGI--LN-IGTIEDYALSEWQRILDIN-VTGVFLGIRAAVKPMKEA
MAV_2509|M.avium_104 LVNNAGI--IN-IGTLEDYALSEWQRILDIN-LTGVFLGIRAVVKPMKKA
MUL_1037|M.ulcerans_Agy99 LVNNVGI--VA-LGQLKKFDLGKWQKVIDVN-LTGTFLGMRAAVEPMTAA
MSMEG_0283|M.smegmatis_MC2_155 LVHVAGGSSSP-AGGFAALSEQDWIDELNLNLLSAVRL-DRAVAPMMIEA
MAB_0607|M.abscessus_ATCC_1997 VVNNAGG--AP-FALAADASHNFHRKIIELNLLSALLVSQRANAVMQKQD
Mflv_0943|M.gilvum_PYR-GCK VVNNAGATTLS-LGGVLGITDADWRKDLDINFLAAVRLNAALLPAMYTR-
Mvan_5918|M.vanbaalenii_PYR-1 IVNNAGATVPH-LGGVLDIEDAEWVNDLNINFLAAVRLNAALLPALYAR-
MLBr_01807|M.leprae_Br4923 LVANAGITSD---MLIMRMSEEQFQKVIDVN-LTGVFRVAKLASRNMIKQ
TH_3911|M.thermoresistible__bu LVNNVGGGSSNEFGYIHDLDPDAWLDSFNRN-VMSAIRMTRAVVPDMLAA
:* .* :: * : .
Mb2025|M.bovis_AF2122/97 GRGSIINISSIEGLAGT-VACHGYTATKFAVRGLTKSTALELGPSGIRVN
Rv2002|M.tuberculosis_H37Rv GRGSIINISSIEGLAGT-VACHGYTATKFAVRGLTKSTALELGPSGIRVN
MMAR_2981|M.marinum_M GRGSIINISSIEGLAGT-IASHGYTVSKFAVRGLTKSTALELGPSGIRVN
MAV_2509|M.avium_104 GRGSIINISSIEGMAGT-IACHGYTATKFAVRGLTKSAALELGPSGIRVN
MUL_1037|M.ulcerans_Agy99 GSGSIINVSSIEGLRGA-PAVHPYVASKWAVRGLTKSAALELAPLNIRVN
MSMEG_0283|M.smegmatis_MC2_155 GGGVIVHISSIQSRMPLYDGTLGYAAAKAALRTYSKGLANELAPKGIRVN
MAB_0607|M.abscessus_ATCC_1997 GGGTIVNITSVSAGRPS-PGTAAYGAAKAGLESLTRSLAVEWAPK-VRVN
Mflv_0943|M.gilvum_PYR-GCK GAGSIVNVSSTVILSPP-APMLHYATAKAALATYTTGLAAEAGPRGVRVN
Mvan_5918|M.vanbaalenii_PYR-1 GKGSIINVSSAVTLSPP-SPMLHYATAKAALATYTTGLAAEAGPRGVRVN
MLBr_01807|M.leprae_Br4923 RFGRYIFMGSVVG-LSGVPGQANYAASKSGLIGMARSLARELGSRSITAN
TH_3911|M.thermoresistible__bu SWGRIVMMSSLEGKMPTLPKIGPYVTSKHALLGLTKSLAFDYGAHGITCN
* : : * * .:* .: : . * : .. : *
Mb2025|M.bovis_AF2122/97 SIHPGLVKTPMTDWVPEDIFQTA--------------------LGRAAEP
Rv2002|M.tuberculosis_H37Rv SIHPGLVKTPMTDWVPEDIFQTA--------------------LGRAAEP
MMAR_2981|M.marinum_M SIHPGLVKTPMTEWVPEDLFQTA--------------------LGRAAEP
MAV_2509|M.avium_104 SIHPGLIKTPMTEWVPEDIFQSA--------------------LGRAAEP
MUL_1037|M.ulcerans_Agy99 SIHPGFIRTPMTANLPDDMVTIP--------------------LGRPAES
MSMEG_0283|M.smegmatis_MC2_155 TVSPGFIRTTAAEALIERIAEAGNGTREQALDSLMASLGG-IPLGRPAEP
MAB_0607|M.abscessus_ATCC_1997 ALAAGMIRTELSEMHYG--DEDG-----------IAAVGATVPLGRLANP
Mflv_0943|M.gilvum_PYR-GCK TVTPGNVTSPGADAIRQAIVDSGGADA-------PAAGGTPIPLGRKGEA
Mvan_5918|M.vanbaalenii_PYR-1 TLTPGTVGSPGADAIRRAIIDGVGADR-------PEAGGIPIPLGRKGEA
MLBr_01807|M.leprae_Br4923 VVAPGFIDTEMTRAMDSKYQEAALQAIP------------LGRIG---AV
TH_3911|M.thermoresistible__bu AVCPGYVHIPTRQRKASKAFDEGRFADPQ------QNYKDLSRIGRHIEL
: .* : :*
Mb2025|M.bovis_AF2122/97 VEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWV
Rv2002|M.tuberculosis_H37Rv VEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWV
MMAR_2981|M.marinum_M MEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHKDFSAVDVAAQPEWV
MAV_2509|M.avium_104 KEVSNLVVYLASDESSYSTGSEFVVDGGTTAGLGHKDFSNVETDAQPDWV
MUL_1037|M.ulcerans_Agy99 REVSTFVVFLASDDASYATGSEFVMDGGLVTDVPHKQF------------
MSMEG_0283|M.smegmatis_MC2_155 DEVAGVVGFLVSDAAASVVGADIVVDGG-TVPAI----------------
MAB_0607|M.abscessus_ATCC_1997 DEIGWTAAFLASPLSSYVTGSVLTVHGGGERPAFLDAANVNSNENGEKE-
Mflv_0943|M.gilvum_PYR-GCK SDIAEAIAFLVSDRAAWITATNLVIDGGQIQAG-----------------
Mvan_5918|M.vanbaalenii_PYR-1 SDIAEAIAFLASDRAAWITGSNLIVDGGQIQTR-----------------
MLBr_01807|M.leprae_Br4923 EEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-----------------
TH_3911|M.thermoresistible__bu DEVAHAVSMLVAEESSAITGTTLNIDGGSSPY------------------
::. *.: :. .: : :.**
Mb2025|M.bovis_AF2122/97 T
Rv2002|M.tuberculosis_H37Rv T
MMAR_2981|M.marinum_M T
MAV_2509|M.avium_104 T
MUL_1037|M.ulcerans_Agy99 -
MSMEG_0283|M.smegmatis_MC2_155 -
MAB_0607|M.abscessus_ATCC_1997 -
Mflv_0943|M.gilvum_PYR-GCK -
Mvan_5918|M.vanbaalenii_PYR-1 -
MLBr_01807|M.leprae_Br4923 -
TH_3911|M.thermoresistible__bu -