For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAERLAGKVALVSGGARGMGASHVRSLVAEGAKVVFGDILDDEGKAVAAEVGEATRYLHLDVTKPEDWDA AVATALAEFGRIDVLVNNAGIINIGTLEDYALSEWQRILDINLTGVFLGIRAVVKPMKKAGRGSIINISS IEGMAGTIACHGYTATKFAVRGLTKSAALELGPSGIRVNSIHPGLIKTPMTEWVPEDIFQSALGRAAEPK EVSNLVVYLASDESSYSTGSEFVVDGGTTAGLGHKDFSNVETDAQPDWVT
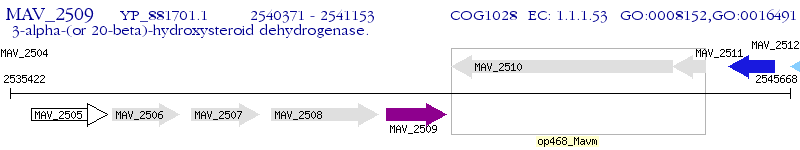
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2509 | - | - | 100% (260) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_5136 | - | 2e-89 | 63.52% (244) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_1572 | - | 5e-45 | 38.33% (240) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 1e-124 | 83.66% (257) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4505 | - | 1e-118 | 78.60% (257) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 1e-124 | 83.66% (257) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-26 | 32.64% (239) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 3e-60 | 49.17% (242) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2981 | fabG3_1 | 1e-124 | 83.85% (260) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 2e-87 | 64.05% (242) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_3693 | - | 3e-47 | 41.25% (240) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 3e-91 | 65.16% (244) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 3e-82 | 58.94% (246) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 ----------MSGR----LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv ----------MSGR----LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
MMAR_2981|M.marinum_M ----------MAGR----LTGKVALVSGGARGMGASHVRALVAEGAHVVL
MAV_2509|M.avium_104 ----------MAER----LAGKVALVSGGARGMGASHVRSLVAEGAKVVF
Mflv_4505|M.gilvum_PYR-GCK ----------MTGR----LAGKVALISGAARGMGASHARVMAGHGAKVVC
MSMEG_3515|M.smegmatis_MC2_155 -----------MGR----VAGKVALISGGARGMGAAHARALVAEGAKVVI
Mvan_3000|M.vanbaalenii_PYR-1 -----------MGR----VDGKVALISGGAQGMGAADARALIAEGAKVVI
MUL_1037|M.ulcerans_Agy99 -----------MGR----VDGKVALISGGARGMGASHARLLVQEGAKVVI
MAB_4680c|M.abscessus_ATCC_199 ----------MNGNS-ALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_3693|M.thermoresistible__bu -----------VSL----LAGQTAVVTGGARGLGYAIAERFIAEGARVVI
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. .: :::*. :*:* : . : .* :*.
Mb2025|M.bovis_AF2122/97 GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
Rv2002|M.tuberculosis_H37Rv GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
MMAR_2981|M.marinum_M GDILDDEGRAVAAELG----DAARYVHLDVTQPEQWTAAVDTAVNEFGGL
MAV_2509|M.avium_104 GDILDDEGKAVAAEVG----EATRYLHLDVTKPEDWDAAVATALAEFGRI
Mflv_4505|M.gilvum_PYR-GCK GDILDSDGEAVAAELG----DAARYVHLDVTSPDDWDRAVAAAVADFGGL
MSMEG_3515|M.smegmatis_MC2_155 GDILDDEGAALAAELG----DAARYVHLDVTQAEDWDTAVATATTDFGLL
Mvan_3000|M.vanbaalenii_PYR-1 GDILDDKGKALADEINAETPDSIRYVHLDVTQADQWEAAVATAVDAFGKL
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIG----DAARYVHLDVTQPDQWEAAVATAVDEFGKL
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELG----DAARFTHLDVTDAEQWRELIDTTLAGFGTI
TH_3693|M.thermoresistible__bu GDIDESATRAAADSLGG--ADVAVGMRCDVTRSDEVEALVGAAVKRFGSL
MLBr_01807|M.leprae_Br4923 THRASVVP------------DWFFGVECDVTDNDAIGAAFKAVEEHQGPV
. . : . *** . :. * :
Mb2025|M.bovis_AF2122/97 HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
Rv2002|M.tuberculosis_H37Rv HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
MMAR_2981|M.marinum_M HVLVNNAGILNIGTIEDYALSEWQRILDINVTGVFLGIRAAVKPMKEAGR
MAV_2509|M.avium_104 DVLVNNAGIINIGTLEDYALSEWQRILDINLTGVFLGIRAVVKPMKKAGR
Mflv_4505|M.gilvum_PYR-GCK DVLVNNAGILNIGTVEDYELSEWHRILDVNLTGVFLGIRAATPTMKAAGR
MSMEG_3515|M.smegmatis_MC2_155 NVLINNAGIVALGAIGKFDMTKWQNVIDVNLTGTFLGMQASVGAMKAAGG
Mvan_3000|M.vanbaalenii_PYR-1 NVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKTAGG
MUL_1037|M.ulcerans_Agy99 DVLVNNVGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAGS
MAB_4680c|M.abscessus_ATCC_199 TGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGC
TH_3693|M.thermoresistible__bu DVMVNNAGITRDATMRKMTEQQFDEVIAVHLKGTWNGLRAAAAIMREQKR
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQRF
:: *.* : .: .:: ::: *.: : *
Mb2025|M.bovis_AF2122/97 GSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIH
Rv2002|M.tuberculosis_H37Rv GSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIH
MMAR_2981|M.marinum_M GSIINISSIEGLAGTIASHGYTVSKFAVRGLTKSTALELGPSGIRVNSIH
MAV_2509|M.avium_104 GSIINISSIEGMAGTIACHGYTATKFAVRGLTKSAALELGPSGIRVNSIH
Mflv_4505|M.gilvum_PYR-GCK GSIINIASIEGMAGTVGCHGYTATKFAVRGLTKSTALELGPFGIRVNSVH
MSMEG_3515|M.smegmatis_MC2_155 GSIINVSSIEGLRGAPMVHPYVASKWAVRGLTKSAALELGAHQIRVNSIH
Mvan_3000|M.vanbaalenii_PYR-1 GSIINISSIEGLRGAVMVHPYVASKWAVRGLTKSAALELGSHNIRVNSVH
MUL_1037|M.ulcerans_Agy99 GSIINVSSIEGLRGAPAVHPYVASKWAVRGLTKSAALELAPLNIRVNSIH
MAB_4680c|M.abscessus_ATCC_199 GSIVNISSAAGLVGLALTSGYGASKWGVRGLSKVAAIELGRERIRVNSVH
TH_3693|M.thermoresistible__bu GAIVNMSSISGKVGMVGQTNYSAAKAGIVGMTKAASKELAYLGVRVNAIQ
MLBr_01807|M.leprae_Br4923 GRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVVA
* : :.* * * * .:* .: *::: : **. : .* :
Mb2025|M.bovis_AF2122/97 PGLVKTPMTDWV-----PEDIFQTALGRAAEPVEVSNLVVYLASDESSYS
Rv2002|M.tuberculosis_H37Rv PGLVKTPMTDWV-----PEDIFQTALGRAAEPVEVSNLVVYLASDESSYS
MMAR_2981|M.marinum_M PGLVKTPMTEWV-----PEDLFQTALGRAAEPMEVSNLVVYLASDESSYS
MAV_2509|M.avium_104 PGLIKTPMTEWV-----PEDIFQSALGRAAEPKEVSNLVVYLASDESSYS
Mflv_4505|M.gilvum_PYR-GCK PGLVKTPMADWV-----PEDIFQSALGRIAQPHEVSNLVVYLASDESSYS
MSMEG_3515|M.smegmatis_MC2_155 PGFIRTPMTEHF-----PDDMLRIPLGRPGEVDEVSSFVVFLASDESRYA
Mvan_3000|M.vanbaalenii_PYR-1 PGFIRTPMTKHF-----PDNMLRIPLGRPGQPEEVATFVVFLASDESRYA
MUL_1037|M.ulcerans_Agy99 PGFIRTPMTANL-----PDDMVTIPLGRPAESREVSTFVVFLASDDASYA
MAB_4680c|M.abscessus_ATCC_199 PGVIYTPMTAKEGFREGEGNMPIAAMGRVGNADEVAGAVAYLLSDAASYV
TH_3693|M.thermoresistible__bu PGLIRSAMTEAMPQRIWDEKLAEIPMGRAGEPEEVAKVALFLASDLSSYM
MLBr_01807|M.leprae_Br4923 PGFIDTEMTRAMDSKYQEAALQAIPLGRIGAVEEIAGAVSFLASEDAGYI
**.: : *: : .:** . *:: . :* *: : *
Mb2025|M.bovis_AF2122/97 TGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv TGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
MMAR_2981|M.marinum_M TGAEFVVDGGTVAGLAHKDFSAVDVAAQPEWVT
MAV_2509|M.avium_104 TGSEFVVDGGTTAGLGHKDFSNVETDAQPDWVT
Mflv_4505|M.gilvum_PYR-GCK TGAEFVVDGGTLAGLAHKDFSAVDVDQQPEWIT
MSMEG_3515|M.smegmatis_MC2_155 TGAEFVMDGGLVNDVPHKL--------------
Mvan_3000|M.vanbaalenii_PYR-1 TGAEFVMDGGLTNDVPHK---------------
MUL_1037|M.ulcerans_Agy99 TGSEFVMDGGLVTDVPHKQF-------------
MAB_4680c|M.abscessus_ATCC_199 TGAELAVDGGWTAGPTLTTITGQ----------
TH_3693|M.thermoresistible__bu TGTVLEVTGGRHL--------------------
MLBr_01807|M.leprae_Br4923 SGAVIPVDGGLGMGH------------------
:*: : : **