For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LPDLSVNRSTTVERVSDALRAAVLSGELTPGTPLREVDLAERMKVSRGSVREALVRLSDEGLLRRTSFRG VEVKQLTADEIRDLFVVRKLIELTAVDAAGTAGAAALAELRRAVDEFGQAIRTGAAVDRNRADMFVHITL VGLLNSPRLSRIHGELMSELQLALVSQYRDSTILPGTELTNRHEEFLRMLLDGDTVGAREQLERRLDLAQ RLLLGTADPGEN*
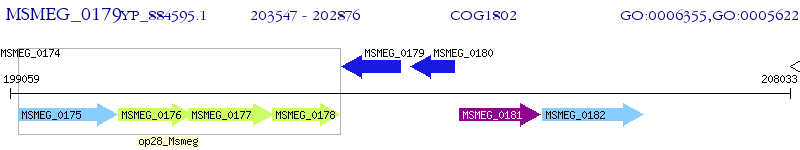
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0179 | - | - | 100% (223) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_2489 | - | 2e-19 | 34.59% (159) | transcriptional regulator, GntR family protein, putative |
| M. smegmatis MC2 155 | MSMEG_4057 | - | 6e-14 | 27.72% (202) | transcriptional regulator, GntR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0170c | - | 3e-09 | 33.87% (124) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0958 | - | 1e-10 | 25.60% (207) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0165c | - | 3e-09 | 33.87% (124) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4411c | - | 4e-21 | 32.88% (219) | GntR family transcriptional regulator |
| M. marinum M | MMAR_2244 | - | 6e-19 | 27.70% (213) | GntR family transcriptional regulator |
| M. avium 104 | MAV_5020 | - | 1e-10 | 35.77% (123) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_0156 | - | 4e-11 | 30.37% (214) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_1833 | - | 5e-19 | 27.70% (213) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0931 | - | 4e-16 | 34.56% (217) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0170c|M.bovis_AF2122/97 MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
Rv0165c|M.tuberculosis_H37Rv MIKHDVVWVTLWPERPNNKPPPSPRQVPGNPGPTLKVLASHVNAPLSAKP
MAV_5020|M.avium_104 ---------------------------------------------MSMQP
TH_0156|M.thermoresistible__bu -----------------------------------------MNAPITSRL
MSMEG_0179|M.smegmatis_MC2_155 ------------------------------------------MLPDLS--
MAB_4411c|M.abscessus_ATCC_199 --------------------------------------MTTLVPEEFAP-
MMAR_2244|M.marinum_M ----------------------------------MLEEEAVVIPRADSPD
MUL_1833|M.ulcerans_Agy99 ----------------------------------MLEEEAVVIPRADSPD
Mflv_0958|M.gilvum_PYR-GCK --------------------------------------------MADAQL
Mvan_0931|M.vanbaalenii_PYR-1 -----------------------------------------MPHRRHSVL
Mb0170c|M.bovis_AF2122/97 RSQLP-LRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTP
Rv0165c|M.tuberculosis_H37Rv RSQLP-LRRAQLSDEVAGHLRAAIMSGALRSGTFIRLDETAAELGVSVTP
MAV_5020|M.avium_104 RSRRP-LRRAQLSDEVAGHLRAAIMSGALRPGTFIRLDETAAELGVSVTP
TH_0156|M.thermoresistible__bu GPNGGGRRRDQLSDEVAAQLRAEIMSGALRPGTYIRLDETAARLGVSITP
MSMEG_0179|M.smegmatis_MC2_155 ------VNRSTTVERVSDALRAAVLSGELTPGTPLREVDLAERMKVSRGS
MAB_4411c|M.abscessus_ATCC_199 ------LERQSTAELIAERLRIAIMRGVLAPGAQLGEASLAAQFAVSRGP
MMAR_2244|M.marinum_M QTCGRLPARGTIGEQVADLIQESILSGEAKRGAPLREELIADRYQVSRRT
MUL_1833|M.ulcerans_Agy99 QTCGRLPARGTVGEQVADLIQESILSGEAKRGSPLREELIADRYQVSRRT
Mflv_0958|M.gilvum_PYR-GCK QLAALPAAEGRRAEQVMAAVRAALDAGVMRPGVKYSVYQLADALQVSRTP
Mvan_0931|M.vanbaalenii_PYR-1 LGSLTVERPGGSQQVILDELRRAILDGAVPPGTPIPLAEVAEHFGVSRIP
: : :: : * * * ** .
Mb0170c|M.bovis_AF2122/97 VREALLKLRGEGMVGLEPHRGHVVLPLTRQDIDDIFWLQATIAQELATSA
Rv0165c|M.tuberculosis_H37Rv VREALLKLRGEGMVGLEPHRGHVVLPLTRQDIDDIFWLQATIAQELATSA
MAV_5020|M.avium_104 VREALLKLRGEGMVQLEPHRGHVVLPLTRQDIEDIFWLQATIAKELAAAA
TH_0156|M.thermoresistible__bu IREALRTLRGEGMVQLEPHRGHVVAPLTRGDIEDIFWLQATIARKLTAAA
MSMEG_0179|M.smegmatis_MC2_155 VREALVRLSDEGLLRRTSFRGVEVKQLTADEIRDLFVVRKLIELTAVDAA
MAB_4411c|M.abscessus_ATCC_199 LREAMQRLVSEGLLRSERNRGIFVIELTDDDVLDVYRSRSAIERAALECI
MMAR_2244|M.marinum_M VRDALRVLESNGLVRHQRHKGSTVVDFSAEDITDMYRSREVLELDAAKRW
MUL_1833|M.ulcerans_Agy99 VRDALRVLESNGLVRHQRHKGSTVVDFSAGDITDMYRSREVLELDAAKRW
Mflv_0958|M.gilvum_PYR-GCK VRDALLRLEEIGLIRFEARQGFRILLPDPREIADIVAIRFALEPPAAGRA
Mvan_0931|M.vanbaalenii_PYR-1 VRESLKTLIGEGLVAHRANSGYMVAQLTAAELGELYVARESLEAASLAAA
:*::: * *:: * : :: :: : :
Mb0170c|M.bovis_AF2122/97 TAHITDVEIDELDRINNALA---GAIGSGDAKTIASIEFAFHRVFNKASR
Rv0165c|M.tuberculosis_H37Rv TAHITDVEIDELDRINNALA---GAIGSGDAKTIASIEFAFHRVFNKASR
MAV_5020|M.avium_104 TDHITDTEIDELDRINDALA---AAVGSGDAETIAGLEFGFHRVFNQASG
TH_0156|M.thermoresistible__bu VDRITDAELDELTALNEALA---AAVDRGVVGEVAAAMSAFHRVLHRTTG
MSMEG_0179|M.smegmatis_MC2_155 GTAG-AAALAELRRAVDEFG---QAIRTGAAVDRNRADMFVHITLVGLLN
MAB_4411c|M.abscessus_ATCC_199 MTGDTVTAYRRLLVPVAVMT---EAAARGDVVAVTDADQDFHEVLVDCSG
MMAR_2244|M.marinum_M CRAGEVDSGYRFHRLTEALGRLEQASRGTDSGLIVKSDLDFHAAIIGLLD
MUL_1833|M.ulcerans_Agy99 CRAGEVDSGYRFHRLTEALGRLEQASRGTDSGLIVKSDLDFHAAIIGLLD
Mflv_0958|M.gilvum_PYR-GCK AAVCGPPLADRLAERMELLR---RAAARGDEPGFAAHDLLLHDHILEAAG
Mvan_0931|M.vanbaalenii_PYR-1 VGNASEADFATAIDMNALLE---HAIREDDAVAYHRQSRRFHAALTGPSR
: * .* :
Mb0170c|M.bovis_AF2122/97 RIKLAWFLLNAARYMPAQVFAADP------RWGADAVNSHRQLIAALRRR
Rv0165c|M.tuberculosis_H37Rv RIKLAWFLLNAARYMGAGVRGRP----------AMGRGRGEQSSAADRR-
MAV_5020|M.avium_104 RIKLAWFLLNAARYMPVLVYAADP------QWGRAAVDNHRQLIAALRRR
TH_0156|M.thermoresistible__bu RIKLAWFLYHVARYMPPQIYATDQ------QWGTATVARHRELIAALRRR
MSMEG_0179|M.smegmatis_MC2_155 SPRLSRIHGELMSELQLALVSQYR--DSTILPGTELTNRHEEFLRMLLDG
MAB_4411c|M.abscessus_ATCC_199 SPRLRRAMRTLLLETRMLLGELQG--AYPDLS--EQVREHEVLCAAIGAG
MMAR_2244|M.marinum_M SPRVDRFFAAIEAEMLYALAILEASEGEYKTDAAKAFGEHKAMYEALRDR
MUL_1833|M.ulcerans_Agy99 SPRVDRFFAAIEAEMLYALAILEASEGEYKTDAAKAFGEHKAMYEALRDR
Mflv_0958|M.gilvum_PYR-GCK NNRARRIVRSLRESTRLLGGITAD----RSRTMSDIDAEHQPVVDAVIAN
Mvan_0931|M.vanbaalenii_PYR-1 MYRLLRMLETAWNLTEPVQPMVHVT----PDDRIRLHADHGEMLQAFLTR
:
Mb0170c|M.bovis_AF2122/97 DTAAVIEHTVWQFTDGARRLTEALAETEVFG------------------
Rv0165c|M.tuberculosis_H37Rv --AAPPRHSRR---NRAHRLAVHRWGTQADGGPG---------------
MAV_5020|M.avium_104 DTAAVIEHTVWQFTDAAARLTAMRDRTGTPSNRG---------------
TH_0156|M.thermoresistible__bu DTETATRVTVDLFTDSAQRLIAQLEKVGIWT------------------
MSMEG_0179|M.smegmatis_MC2_155 DTVGAREQLERRLDLAQRLLLGTADPGEN--------------------
MAB_4411c|M.abscessus_ATCC_199 DAPAAYRLIDEHMHDAVTRLMARRDPGSVE-------------------
MMAR_2244|M.marinum_M DEQRSTQLISEHLRINRDVLIRIVAS-----------------------
MUL_1833|M.ulcerans_Agy99 DEQRSTQLISEHLRINRDVLIRIVAS-----------------------
Mflv_0958|M.gilvum_PYR-GCK DPAAAAAAMRTHLTSTGRLLVAQAVRDQDSPLDAAAVWAEAVGANTELP
Mvan_0931|M.vanbaalenii_PYR-1 DVERLLGAAERHARRLNSVIATLSFETGLLAAEKDISSPQ---------
: