For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSALANYREITDANNRVYRIGESDREILGRSRSWMVWLPWLAMMAVSVYEYGYGAAEEAIRDAHGWSMS ETFWLLAIWALFQAGIAFPAGKLREKGIVSARAAMLIGAALSALGFVTLTQGNLFVAYLGFAVCGGIGAG LVYATCINMVGKWFPERRGGKTGFVNGGFAYGAVPFIFIFSYALHPSTYVWVLDLVGAYMLVVLVICGLM FRDPPKNWWPESVDPLQWANSKAGAKGLAKNPPAVKQFTPMEAIKTGMLPLMWLSLGISAGVSLFGSSYM VPFAKELGFGPLIAASSAGVLSIINGTGRGATGWVSDRLGRKLTLILVLLVSAVALVGLLYAGQAHNEVL FLFFAFLVGFGGGAFYPMFAALTPDYFGENNNASNYGLVYSSKLLGSVVGIGVGASVIDTWGYAGAYWIA AASALVSAAIAVFLRQPGRRSGAAVAPSSTKTSTLASASD
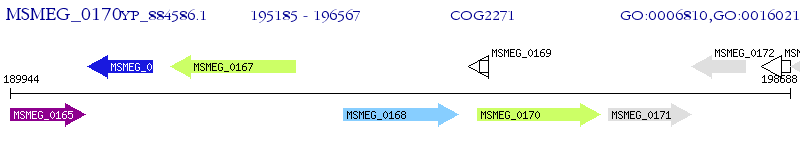
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0170 | - | - | 100% (460) | transmembrane transport protein |
| M. smegmatis MC2 155 | MSMEG_0167 | - | e-151 | 57.89% (437) | transmembrane transport protein |
| M. smegmatis MC2 155 | MSMEG_0164 | - | 1e-94 | 41.03% (429) | transmembrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 6e-06 | 31.97% (122) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_1096 | - | 5e-12 | 24.00% (400) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 6e-06 | 31.97% (122) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_00556 | - | 1e-06 | 31.78% (107) | putative integral membrane drug transport protein |
| M. abscessus ATCC 19977 | MAB_0284c | - | 9e-08 | 26.77% (127) | major facilitator transporter |
| M. marinum M | MMAR_0093 | ermB | 3e-08 | 24.73% (182) | integral membrane efflux protein ErmB |
| M. avium 104 | MAV_3974 | - | 1e-07 | 22.14% (393) | permease of the major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_1008 | - | 4e-07 | 26.49% (151) | AMINOGLYCOSIDES/TETRACYCLINE-TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0354 | - | 3e-06 | 25.81% (186) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_2691 | - | 7e-08 | 27.04% (196) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0805c|M.bovis_AF2122/97 MLGNAMVEACPAEGDAP-VPITPAGRPRSGQRSYPDRLDVGLLRTAGVCV
Rv0783c|M.tuberculosis_H37Rv MLGNAMVEACPAEGDAP-VPITPAGRPRSGQRSYPDRLDVGLLRTAGVCV
MMAR_0093|M.marinum_M -MNTTPAKATALQQNLPGRPDGSTGSSVAAGERHVDRLDGSLLRIAGICV
TH_1008|M.thermoresistible__bu -----MRRATAAAIRPPRG----HNRTIAISAGSLAVLLGALDTYVVITI
Mvan_2691|M.vanbaalenii_PYR-1 -----MHRTPITAKPAPHAGRGAINRRIAISAGSLAVVLGALDTYVVVTI
MLBr_00556|M.leprae_Br4923 -----MSTRAG--------------RRVAISAGSLAVLLGALDTYVVVTI
MAB_0284c|M.abscessus_ATCC_199 ------------------------------MTRVVLIVLCALQVTMQLYV
Mflv_1096|M.gilvum_PYR-GCK -------MGGPVTRFDSAAIAEWRRYGTVPLAAGLGYSMGVIHVYSLGAF
MAV_3974|M.avium_104 --------------------------------------MVALGYGVISPA
MSMEG_0170|M.smegmatis_MC2_155 ------MSSALANYREITDANNRVYRIGESDREILGRSRSWMVWLPWLAM
MUL_0354|M.ulcerans_Agy99 MLTSGSMTTTATIAAGEAMRVSSSSSPDLPDAGDTTKFLRPRNAALRSKA
Mb0805c|M.bovis_AF2122/97 LASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTGYMLALATV-----
Rv0783c|M.tuberculosis_H37Rv LASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTGYMLALATV-----
MMAR_0093|M.marinum_M FASVAVNLANTAVSVAQRSLIVTFGSNQAVVAWTVTAYTLTEAAA-----
TH_1008|M.thermoresistible__bu MVDIMRDVGIALNKIHQ-------------VTPIITGYLLGYIAA-----
Mvan_2691|M.vanbaalenii_PYR-1 MVDIMRDVGIGVNEIQR-------------VTPIITGYLLGYIAA-----
MLBr_00556|M.leprae_Br4923 MRDIMHDVGIPVNQMQR-------------ITWIVTMYLLGYIAA-----
MAB_0284c|M.abscessus_ATCC_199 WIPIHDIVSASYGGQDM--------------TPALSAALFGYAAG-----
Mflv_1096|M.gilvum_PYR-GCK MGPLQDEFGWSRAQASVG----------------LTIVGMAAAVA-----
MAV_3974|M.avium_104 LPTFARSFGVSIKAVTF----------------LVTVFSLSRLCF-----
MSMEG_0170|M.smegmatis_MC2_155 MAVSVYEYGYGAAEEAIRDAHG---WSMSETFWLLAIWALFQAGI-----
MUL_0354|M.ulcerans_Agy99 RGGSRSAARMANTKTNRKPRSARALCARAVAAPLGGARPLGERAGGRHRP
:
Mb0805c|M.bovis_AF2122/97 ----------IPTAGWAADR-------FGTRRLFMGSVLAFTLGSLLCAV
Rv0783c|M.tuberculosis_H37Rv ----------IPTAGWAADR-------FGTRRLFMGSVLAFTLGSLLCAV
MMAR_0093|M.marinum_M ----------IPLSGWAADR-------IGTKRLFMISVLGFTLGSVLCAV
TH_1008|M.thermoresistible__bu ----------MPLLGRASDR-------FGRKMLIQVGLAGFAIGSVVTAL
Mvan_2691|M.vanbaalenii_PYR-1 ----------MPLLGRASDR-------FGRKILIQVSLAGFAAGSVITAM
MLBr_00556|M.leprae_Br4923 ----------MPLLSRASDR-------FGRKLLLQVSLAGFAIGSVMTAL
MAB_0284c|M.abscessus_ATCC_199 ----------FLLFGPVTDR-------FGRRRVLLAGVAVMAVLTFLVPF
Mflv_1096|M.gilvum_PYR-GCK ----------ALPIGLMVDR-------LGPRRVGIIGAGLMAGAFALLGT
MAV_3974|M.avium_104 ----------APISGLLTER-------LGERRIYIGGLLIVAVSTAACAF
MSMEG_0170|M.smegmatis_MC2_155 ----------AFPAGKLREKGI-----VSARAAMLIGAALSALGFVTLTQ
MUL_0354|M.ulcerans_Agy99 AREGYPAVIWLLLGGNLVVRAAGFAYPFMAFHVAGRGFAAAAVGAVLAAF
. : . . :
Mb0805c|M.bovis_AF2122/97 AP---NILLLIIFRVVQGFGGGMLTPVSFAILAREAGPKRLGRVMAVVGI
Rv0783c|M.tuberculosis_H37Rv AP---NILLLIIFRVVQGFGGGMLTPVSFAILAREAGPKRLGRVMAVVGI
MMAR_0093|M.marinum_M AP---NIACLIIFRAVQGGGGGILMPLVITILAREAGPNRLARLMSVMGI
TH_1008|M.thermoresistible__bu AGDLQSLHMLVTGRIIQGAASGALLPVTLALAADLWAERNRAGVLGGIGA
Mvan_2691|M.vanbaalenii_PYR-1 S---TDLVVLVVGRVIQGTASGALLPVTLALAADLWAARNRAAVLGGVGA
MLBr_00556|M.leprae_Br4923 AGQFGDFHMLIAGRTIQGVASGALLPITLALGADLWAQRNRAGVLGGIGA
MAB_0284c|M.abscessus_ATCC_199 AP---TPVLSGLFRVIQGFAGGAFSPSALSYLGEALPRRAATTAIGAVST
Mflv_1096|M.gilvum_PYR-GCK ATG--TMGNWILLWSVLAFACFWVQTTVWTSAVASRFETSRGLAFAVTLS
MAV_3974|M.avium_104 SQ---AYWQLMLCRVFSGVGSTMFYVSALGLMIHISPADARGRIAGLFTT
MSMEG_0170|M.smegmatis_MC2_155 GN---LFVAYLGFAVCGGIGAGLVYATCINMVGKWFPER-RGGKTGFVNG
MUL_0354|M.ulcerans_Agy99 GVG-WAVGQLVCGWLVDSLGPRATLAWTMTIAAGVLVGMAEARSVAALLV
. . . .
Mb0805c|M.bovis_AF2122/97 PMLLGPVGGPILGGWLIGAYG-WRWIFLVNLPVGLSALVLAAIVFP-RDR
Rv0783c|M.tuberculosis_H37Rv PMLLGPVGGPILGGWLIGAYG-WRWIFLVNLPVGLSALVLAAIVFP-RDR
MMAR_0093|M.marinum_M PLLLGPMAGPILGGWLIDDYG-WQWIFWINVPIGLITVALAAIAFP-GDH
TH_1008|M.thermoresistible__bu AQELGAVLGPLYGIAMVWLFGAWESVFWVNVPLAALAMVMIHFSLPARDP
Mvan_2691|M.vanbaalenii_PYR-1 AQELGSVLGPVYGISLVWLFGHWQSVFWVNVPLAVIAMVMIHFSLPGRSP
MLBr_00556|M.leprae_Br4923 AQELGSVLGPLYGIFIVWLFSDWRYVFWINIPLTAIAMLMIQVSLSAHDR
MAB_0284c|M.abscessus_ATCC_199 AYLMAGVLGQVMASVITQAWG-WQWLFWLSAAGLAVNVALLALVMHEERA
Mflv_1096|M.gilvum_PYR-GCK GGSLAAAVFPPLATLLIGEWGWRGALFGLGGLWGGLVLIVVLLFFRGAQD
MAV_3974|M.avium_104 SFMVGAVGGPAVGG-LAAGWGLTAPFVVYGVAMLGVALVLFLGLRNSALA
MSMEG_0170|M.smegmatis_MC2_155 GFAYGAVPFIFIFSYALHPST-YVWVLDLVGAYMLVVLVICGLMFRDPPK
MUL_0354|M.ulcerans_Agy99 GAMITGVVYDAPRPVMGAAIAELIPEPSRRAKLDAWRFGWIVSIGRAITG
.
Mb0805c|M.bovis_AF2122/97 PAASENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLAL
Rv0783c|M.tuberculosis_H37Rv PAASENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLAL
MMAR_0093|M.marinum_M TAPSETLDIIGMLLLSPGLATFLYGLSTVPARGTVADRHVLIPATAGLVL
TH_1008|M.thermoresistible__bu DGPRERIDVVGGVLLGITLFLIVVGLYNPQPSAREVVPTWRLWLLAAAAV
Mvan_2691|M.vanbaalenii_PYR-1 TEDPQKVDVVGGLLLAVALGLAVIGLYNPKPDGKTVLPSWGLPVLIAAAV
MLBr_00556|M.leprae_Br4923 GDELEKVDVVGGVLLAIALGLVVIGLYNPQPDSKQVLPSYGVPVLVGGIV
MAB_0284c|M.abscessus_ATCC_199 GDPS-----------------------TRVVSGFAAQARLLARADIAVPL
Mflv_1096|M.gilvum_PYR-GCK DKKPAPAR----------------TVIAPTAAAADLPGVTLAEGLRSATF
MAV_3974|M.avium_104 APPP------------------------PTRSTVTMREALRVRAYQSALL
MSMEG_0170|M.smegmatis_MC2_155 NWWPESVDPLQWAN-------SKAGAKGLAKNPPAVKQFTPMEAIKTGML
MUL_0354|M.ulcerans_Agy99 GVGGLLAAWSG------VPVLFWINAVACALLALVAARCIPATAHRPPVA
Mb0805c|M.bovis_AF2122/97 IAAFVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPS
Rv0783c|M.tuberculosis_H37Rv IAAFVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPS
MMAR_0093|M.marinum_M MGAFVFHALYRADRPLIDLRLFRNRVVTVANATIVFVAAGFSGAVLLVPS
TH_1008|M.thermoresistible__bu TLIGFFVWERRARTKLIDPAGVRFRPFLAALGASFVAGAALMVTLVNVEL
Mvan_2691|M.vanbaalenii_PYR-1 AAVAFFGWEKFARTRLLEPRGVHFRPFLAALGASLTAGAALMVTLVNVEL
MLBr_00556|M.leprae_Br4923 ATVAFAVWERCARTRLIDPAGVHIRPFLSALGASVAAGAALMVTLVNVEL
MAB_0284c|M.abscessus_ATCC_199 VAVFVLMLSFVAMYTSLATELVDALHLPESMGTTIRLAG-----------
Mflv_1096|M.gilvum_PYR-GCK YKLLVAAGLFAFTTLGIVVHLVPILRDAGAEPLAAAGTAALVG-------
MAV_3974|M.avium_104 SNFATGWSAFGLRMALVP--LFVSDVMGRGIGTIGVVLAAFAG-------
MSMEG_0170|M.smegmatis_MC2_155 PLMWLSLGISAGVSLFGSSYMVPFAKELGFGPLIAASSAGVLS-------
MUL_0354|M.ulcerans_Agy99 PAVHVRAAHNSYRRAFSDGRLVLLFASSLATLTAVRGLYATVPMLMADSG
.
Mb0805c|M.bovis_AF2122/97 YLQQVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIML
Rv0783c|M.tuberculosis_H37Rv YLQQVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIML
MMAR_0093|M.marinum_M YFQQLLRETPLQVGIHMIPLGLGAAVTIPTSSVLMDRHGAGKVVLGGVTL
TH_1008|M.thermoresistible__bu FGQGVLGKDQFEAAMLLLRFLIALPIGALIGGWLATRVGDRIVTFVGLLI
Mvan_2691|M.vanbaalenii_PYR-1 FGQGVLGQDQNEAAFLLLRFLIALPVGALLGGWLATRVGDRLVTFAGLLI
MLBr_00556|M.leprae_Br4923 FGQGVLGMDQTQAAGLLVWFLIALPIGAVLGGWGATKAGDRTMTFVGLLI
MAB_0284c|M.abscessus_ATCC_199 ---------------------VPGMLVAPFVGSWVGRVGTETGALVGYLL
Mflv_1096|M.gilvum_PYR-GCK ---------------------IFSIVGRLGVGLLLDRFPGRVVGACAYLV
MAV_3974|M.avium_104 ----------------------GNALAVVPSGYLSDRMGRRTLLIVGLVT
MSMEG_0170|M.smegmatis_MC2_155 ---------------------IINGTGRGATGWVSDRLGRKLTLILVLLV
MUL_0354|M.ulcerans_Agy99 LGAGEFGWAQVAN------AAAGIGLTPVITPWLGRKVAARINPRLDILA
:
Mb0805c|M.bovis_AF2122/97 IAAGLGTFAFGVARQADYL---------PILPTGLAIMGMGMGCSMMPLS
Rv0783c|M.tuberculosis_H37Rv IAAGLGTFAFGVARQADYL---------PILPTGLAIMGMGMGCSMMPLS
MMAR_0093|M.marinum_M ISVGMGTLAFGAAEHAAYV---------PTLLIGLTIVGMGIGSIMLQLT
TH_1008|M.thermoresistible__bu AAGGFWLISHWPVDLLAAR-HDLGVVTLPVLDTDLAIVGIGLGLVIGPLT
Mvan_2691|M.vanbaalenii_PYR-1 AAGGFLLISKWPVDLLAAR-HDLGFVSLPVLDTDLAIAGLGLGLVIGPLT
MLBr_00556|M.leprae_Br4923 TAGGYWLISHWPVDLLNYRRSIFGLFSVPTMYADLLVAGLGLGLVIGPLS
MAB_0284c|M.abscessus_ATCC_199 AAAGLAVEALGAG-------------SVPIIIAGSNIFVAGIAVSSTAMV
Mflv_1096|M.gilvum_PYR-GCK PILGCAALLIDGSSPLSQT-------------VAAALFGLTLGAEVDVIA
MAV_3974|M.avium_104 SGAATVWLGFVAS--------------LPVFLVAAGVVGVVTGIYMSPLQ
MSMEG_0170|M.smegmatis_MC2_155 SAVALVGLLYAGQAHNEVL-----------FLFFAFLVGFGGGAFYPMFA
MUL_0354|M.ulcerans_Agy99 IAGGWTALTMAGAALAHTT---------FGFAIAVAASAAGEIAWFVIAV
.
Mb0805c|M.bovis_AF2122/97 GAAVQTLAPHQIARGSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIAT
Rv0783c|M.tuberculosis_H37Rv GAAVQTLAPHQIARGSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIAT
MMAR_0093|M.marinum_M TVAVQTLAPHQIARGSTLVSVNQQLSASASTALMSVILTSQFNRSANIAA
TH_1008|M.thermoresistible__bu SAALRVVPAAEHGIASAAVVVARMIGMVIGLAALSAWGLYRFNQHLQSLA
Mvan_2691|M.vanbaalenii_PYR-1 SATLRVVPAAQHGIASAAVVVARMIGMLIGIAALSAWGLYRFNQHLASLP
MLBr_00556|M.leprae_Br4923 SATLRVVPTAQHGIASAAVVVARMTGMLIGVAALTAWGLYRFNQILAGLS
MAB_0284c|M.abscessus_ATCC_199 TMFGDRAAP-RRGAGTALYGFTVFLGASSAPYLAHALG---FRPLLGILV
Mflv_1096|M.gilvum_PYR-GCK YLASRYFGLRNFGGLFGGLVAALSLGTAFGPLAAGATSDHFGGYTNFLIL
MAV_3974|M.avium_104 AAVADILGNEARAGLPVATVQMMSDLGAIVGSMAVGWAAEQIGYGWGFFI
MSMEG_0170|M.smegmatis_MC2_155 ALTPDYFGENNNASNYGLVYSSKLLGSVVGIGVGASVIDTWGYAGAYWIA
MUL_0354|M.ulcerans_Agy99 GTVHRIAPPASGGRYHGIWSMTLAIASVVAPIVSSCSLTHGGHRLVALVT
.
Mb0805c|M.bovis_AF2122/97 AKKVALTPESGAGRGAAVDPSSLPRQTN---FAAQLLHDLSHAYAVVFVI
Rv0783c|M.tuberculosis_H37Rv AKKVALTPESGAGRGAAVDPSSLPRQTN---FAAQLLHDLSHAYAVVFVI
MMAR_0093|M.marinum_M ANRLEALNESATRRGIPLDPSQIPPRTLNPDFGANLLHDLSQAYTVVFVY
TH_1008|M.thermoresistible__bu AREPGGDSLAARLAAEAARYREAYAMQYGEIFAITALLCVAGALLGLLIA
Mvan_2691|M.vanbaalenii_PYR-1 ASTAGG-SLAERLAAEANRVREAYVLQYGEIFTITAVVCLVGALLGLLIA
MLBr_00556|M.leprae_Br4923 TAVPPDATLIERAAAVADQAKRAYTMMYSDIFMITAIVCVIGALLGLLIS
MAB_0284c|M.abscessus_ATCC_199 AALIGGAALVASSRRASG--------------PVKPVVDISTQSSPLP--
Mflv_1096|M.gilvum_PYR-GCK TMVLMGLSSMALASLKPP--------------PVWTVTSPGEDARADRFP
MAV_3974|M.avium_104 SGVVLLIAAVGWVMAPETR------------TATELEADLMAASRTSSRC
MSMEG_0170|M.smegmatis_MC2_155 AASALVSAAIAVFLRQPGR-------------RSGAAVAPSSTKTSTLAS
MUL_0354|M.ulcerans_Agy99 VTVGLAGAALCLPLNRSGD------------------VGVRDSQYTAVAA
Mb0805c|M.bovis_AF2122/97 ATALVVSTLIP-----------------------AAFLPKQQASHRRAPL
Rv0783c|M.tuberculosis_H37Rv ATALVVSTLIP-----------------------AAFLPKQQASHRRAPL
MMAR_0093|M.marinum_M ASVVMLVTFLP-----------------------AVFLP-MRAAITSAPL
TH_1008|M.thermoresistible__bu GHGDRADDTGTPDGSVWDLHGSGDDDTPTRVIGRQVPLPSMEQTVRMPRP
Mvan_2691|M.vanbaalenii_PYR-1 AKDDHAEE------------------------------PSDEAQQRSSPG
MLBr_00556|M.leprae_Br4923 SRKEHASE---------------------------------------PKV
MAB_0284c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1096|M.gilvum_PYR-GCK QP------------------------------------------------
MAV_3974|M.avium_104 EQQL----------------------------------------------
MSMEG_0170|M.smegmatis_MC2_155 ASD-----------------------------------------------
MUL_0354|M.ulcerans_Agy99 AA------------------------------------------------
Mb0805c|M.bovis_AF2122/97 LSA---------------
Rv0783c|M.tuberculosis_H37Rv LSA---------------
MMAR_0093|M.marinum_M REPIDPEFPEMAARGRPG
TH_1008|M.thermoresistible__bu PGD---------------
Mvan_2691|M.vanbaalenii_PYR-1 PIS---------------
MLBr_00556|M.leprae_Br4923 PE----------------
MAB_0284c|M.abscessus_ATCC_199 ------------------
Mflv_1096|M.gilvum_PYR-GCK ------------------
MAV_3974|M.avium_104 ------------------
MSMEG_0170|M.smegmatis_MC2_155 ------------------
MUL_0354|M.ulcerans_Agy99 ------------------