For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRVVLIVLCALQVTMQLYVWIPIHDIVSASYGGQDMTPALSAALFGYAAGFLLFGPVTDRFGRRRVLLAG VAVMAVLTFLVPFAPTPVLSGLFRVIQGFAGGAFSPSALSYLGEALPRRAATTAIGAVSTAYLMAGVLGQ VMASVITQAWGWQWLFWLSAAGLAVNVALLALVMHEERAGDPSTRVVSGFAAQARLLARADIAVPLVAVF VLMLSFVAMYTSLATELVDALHLPESMGTTIRLAGVPGMLVAPFVGSWVGRVGTETGALVGYLLAAAGLA VEALGAGSVPIIIAGSNIFVAGIAVSSTAMVTMFGDRAAPRRGAGTALYGFTVFLGASSAPYLAHALGFR PLLGILVAALIGGAALVASSRRASGPVKPVVDISTQSSPLP*
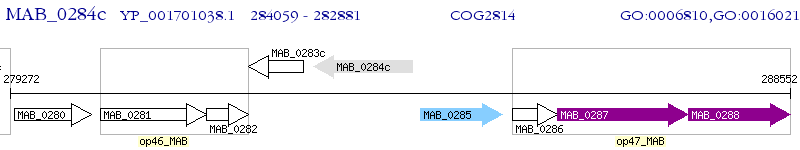
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0284c | - | - | 100% (392) | major facilitator transporter |
| M. abscessus ATCC 19977 | MAB_2706c | - | 1e-20 | 26.24% (362) | putative transporter |
| M. abscessus ATCC 19977 | MAB_2807 | - | 2e-16 | 27.39% (387) | major facilitator transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 4e-17 | 25.50% (353) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_2220 | - | 1e-17 | 25.26% (388) | Bcr/CflA subfamily drug resistance transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 4e-17 | 25.50% (353) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_00556 | - | 4e-11 | 26.79% (321) | putative integral membrane drug transport protein |
| M. marinum M | MMAR_2966 | - | 2e-17 | 23.30% (339) | integral membrane drug efflux protein |
| M. avium 104 | MAV_1539 | - | 5e-19 | 25.79% (349) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_0103 | - | 5e-21 | 26.88% (372) | major facilitator family protein transporter |
| M. thermoresistible (build 8) | TH_1008 | - | 2e-14 | 27.03% (381) | AMINOGLYCOSIDES/TETRACYCLINE-TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0501 | - | 7e-13 | 23.24% (370) | integral membrane drug efflux protein |
| M. vanbaalenii PYR-1 | Mvan_1322 | - | 1e-15 | 25.19% (397) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1539|M.avium_104 --------------------MVMPN-GPVAHQAGTRG---YRRMTAALCG
MSMEG_0103|M.smegmatis_MC2_155 --------------------MVTAQPEPAVWQGHTRGSSDYRRLLAALFC
Mb0805c|M.bovis_AF2122/97 MLGNAMVEACPAEGDAPVPITPAGRPRSGQRSYPDRLDVGLLRTAGVCVL
Rv0783c|M.tuberculosis_H37Rv MLGNAMVEACPAEGDAPVPITPAGRPRSGQRSYPDRLDVGLLRTAGVCVL
MUL_0501|M.ulcerans_Agy99 MLSTAMDEAHCTPAEVAPPDTRTGTSSPGEHVYPDRLDAGLLRIAGVCVL
MMAR_2966|M.marinum_M -----MRRAGTPRRPRSPDTHPDNR---HQHSYPDRLDAGLWRIAGVCVL
MLBr_00556|M.leprae_Br4923 ------------------MSTRAGRR--------VAISAGSL----AVLL
TH_1008|M.thermoresistible__bu ----------------MRRATAAAIRPPRGHNRTIAISAGSL----AVLL
Mvan_1322|M.vanbaalenii_PYR-1 -----------------------------MTDTHRGDDGGTPLALAALLS
MAB_0284c|M.abscessus_ATCC_199 -----------------------------------------MTRVVLIVL
Mflv_2220|M.gilvum_PYR-GCK -------------MSLINFMAISPDVKLKAQSPESAGPPSRTRMIIVLGL
MAV_1539|M.avium_104 AGLASFAAMYCTQALLPALSAHHRIGPATAALTVSLTTGALALSIIPASV
MSMEG_0103|M.smegmatis_MC2_155 AGVATFAQLYSPQAVLPLIASDLGTGAAHAALAISAATIGLAFGVLPWAA
Mb0805c|M.bovis_AF2122/97 ASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTGYMLALATVIPTAGW
Rv0783c|M.tuberculosis_H37Rv ASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTGYMLALATVIPTAGW
MUL_0501|M.ulcerans_Agy99 ASLMSVLDATVVGIAQRTFIIEFDSTQAVVAWTMTGYTLALATVIPVAGW
MMAR_2966|M.marinum_M GSIMTMVDTSVVTVAQRTFVDTFGSTQAVVAWTITGYTLALAAVVPLAGW
MLBr_00556|M.leprae_Br4923 GALDTYVVVTIMRDIMHDVGIPVN-QMQRITWIVTMYLLGYIAAMPLLSR
TH_1008|M.thermoresistible__bu GALDTYVVITIMVDIMRDVGIALN-KIHQVTPIITGYLLGYIAAMPLLGR
Mvan_1322|M.vanbaalenii_PYR-1 GTLVGTISNNVVNVPMPAILADFDAPLESGVFIVVGFLLTFTATMPVVGW
MAB_0284c|M.abscessus_ATCC_199 CALQVTMQLYVWIPIHDIVSASYG--GQDMTPALSAALFGYAAGFLLFGP
Mflv_2220|M.gilvum_PYR-GCK LVALGPLTIDMYLPALPKIGEELSVSSSVAQLTLTGTLAGLAIGQLVIGP
. : .
MAV_1539|M.avium_104 LSERYGRTRVMLISGVASSVIGLLLPFS---PSLGVLVFGRAVQGVALAG
MSMEG_0103|M.smegmatis_MC2_155 LADRVGRVQVITVSVVVATVLGLLVPFA---PTYALLLCGRFVEGLALAG
Mb0805c|M.bovis_AF2122/97 AADRFGTRRLFMGSVLAFTLGSLLCAVA---PNILLLIIFRVVQGFGGGM
Rv0783c|M.tuberculosis_H37Rv AADRFGTRRLFMGSVLAFTLGSLLCAVA---PNILLLIIFRVVQGFGGGM
MUL_0501|M.ulcerans_Agy99 AADRFGTKRLWMGSVLAFALGSLLCALA---PNILSLIFFRVLQGVGGGM
MMAR_2966|M.marinum_M AADRFGTKRMFMGSILVFTLSSLLCAIA---PNIALLIASRVVQGLGGGM
MLBr_00556|M.leprae_Br4923 ASDRFGRKLLLQVSLAGFAIGSVMTALAGQFGDFHMLIAGRTIQGVASGA
TH_1008|M.thermoresistible__bu ASDRFGRKMLIQVGLAGFAIGSVVTALAGDLQSLHMLVTGRIIQGAASGA
Mvan_1322|M.vanbaalenii_PYR-1 IGDRIGRRRVYCASMIGTALCAIGAATA---PTLELLILWRCLGGLAASA
MAB_0284c|M.abscessus_ATCC_199 VTDRFGRRRVLLAGVAVMAVLTFLVPFA---PTPVLSGLFRVIQGFAGGA
Mflv_2220|M.gilvum_PYR-GCK LSDSLGRRKPLFAGIVLHMVASLLCMFA---PTITVLGIARGLQGMGAAA
: * . : . : * : * . .
MAV_1539|M.avium_104 IPAVAMALLAEEVDASSLGSAMGRYIAGTTIGGLAGRIVPSVVVQVGT-W
MSMEG_0103|M.smegmatis_MC2_155 VPAVAVAYLTEEINAGHAARAAGTYVAGTTIGGLAGRLVTGPVAEFAG-W
Mb0805c|M.bovis_AF2122/97 LTPVSFAILAREAGPKRLGRVMAVVGIPMLLGPVGGPILGGWLIGAYG-W
Rv0783c|M.tuberculosis_H37Rv LTPVSFAILAREAGPKRLGRVMAVVGIPMLLGPVGGPILGGWLIGAYG-W
MUL_0501|M.ulcerans_Agy99 LMPLGFIILTRAAGPKRLGRLMAAIGIPMLLGPIGGPILGGWLISSFG-W
MMAR_2966|M.marinum_M LAPLALTIVNREAGPKRVGRVMAVLGIPGVLAPAFGPALGGWLIDSYS-W
MLBr_00556|M.leprae_Br4923 LLPITLALGADLWAQRNRAGVLGGIGAAQELGSVLGPLYGIFIVWLFSDW
TH_1008|M.thermoresistible__bu LLPVTLALAADLWAERNRAGVLGGIGAAQELGAVLGPLYGIAMVWLFGAW
Mvan_1322|M.vanbaalenii_PYR-1 LGPAVMGLLTWLFAGERRGRAVGAWASVNGIGQAIGPTMGGFIADAWG-W
MAB_0284c|M.abscessus_ATCC_199 FSPSALSYLGEALPRRAATTAIGAVSTAYLMAGVLGQVMASVITQAWG-W
Mflv_2220|M.gilvum_PYR-GCK GMVVAIAVVGDLYKDNAAATVMSRLMLVLGVAPVLAPSLGAAVLLHGS-W
. . :. . : *
MAV_1539|M.avium_104 RVALLACSLITLAGTAVFAVLVPR--------------SRFFTPKPASVR
MSMEG_0103|M.smegmatis_MC2_155 RVGVLTVALLCGMAALAFVKLAPA--------------AQGFTPARTHWD
Mb0805c|M.bovis_AF2122/97 RWIFLVNLPVGLSALVLAAIVFPR-DRPAASENFDYMGLLLLSPGLATFL
Rv0783c|M.tuberculosis_H37Rv RWIFLVNLPVGLSALVLAAIVFPR-DRPAASENFDYMGLLLLSPGLATFL
MUL_0501|M.ulcerans_Agy99 HWIFLVNLPIGLAAFALAAITFPA-DRPVPSESFDVIGVLLLSPGLAAFL
MMAR_2966|M.marinum_M QWIFWVNLPVGVVAVGLAAVVFPR-DTPAPSETFDVVGMLLLSPGLPAFL
MLBr_00556|M.leprae_Br4923 RYVFWINIPLTAIAMLMIQVSLSAHDRGDELEKVDVVGGVLLAIALGLVV
TH_1008|M.thermoresistible__bu ESVFWVNVPLAALAMVMIHFSLPARDPDGPRERIDVVGGVLLGITLFLIV
Mvan_1322|M.vanbaalenii_PYR-1 RWVFVPLVPVALAGLIGTLRHIPR--YPGTRMSFDAVGATALTVGSGLLI
MAB_0284c|M.abscessus_ATCC_199 QWLFWLSAAGLAVNVALLALVMHE---------------ERAGDPSTRVV
Mflv_2220|M.gilvum_PYR-GCK HWVFAALVVVAAALLVMAVLALPE--------------TLPVDHRRPLKV
. .
MAV_1539|M.avium_104 AALRN--------------------------------------------L
MSMEG_0103|M.smegmatis_MC2_155 LGRR---------------------------------------------L
Mb0805c|M.bovis_AF2122/97 FGVSS-SPARGTMADRHVLIPAITGLALIAAFVAHSWYRTEHPLIDMRLF
Rv0783c|M.tuberculosis_H37Rv FGVSS-SPARGTMADRHVLIPAITGLALIAAFVAHSWYRTEHPLIDMRLF
MUL_0501|M.ulcerans_Agy99 LALSL-IPDRGTVADRYVLIPVIAGLSLIGGFAWHAWHRADHPLIDLHLF
MMAR_2966|M.marinum_M YGMSE-IPIYGTVADRHVWVPAGIGIALIVGFMFHALYRADKPLIDLRLL
MLBr_00556|M.leprae_Br4923 IGLYNPQPDSKQVLPSYGVPVLVGGIVATVAFAVWER-CARTRLIDPAGV
TH_1008|M.thermoresistible__bu VGLYNPQPSAREVVPTWRLWLLAAAAVTLIGFFVWER-RARTKLIDPAGV
Mvan_1322|M.vanbaalenii_PYR-1 LGVAL----IPRLGVSVAVLSAIAVAVVVLAFFVWHCLRTPEPFVDVRLV
MAB_0284c|M.abscessus_ATCC_199 SGFAA----------------------------------------QARLL
Mflv_2220|M.gilvum_PYR-GCK RGIAA---------------------------------------TYLELV
. .
MAV_1539|M.avium_104 AGHLRNPVLAKLFAVGFVLMGGFVTVYNYLGYRLAARPFGLAPSVVGLLF
MSMEG_0103|M.smegmatis_MC2_155 LTNLRSPRQLALFAQGFLLMGGFVAVYNFLGFRLSAAPFNLPQTVVSLVF
Mb0805c|M.bovis_AF2122/97 QNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQSPMQSGVHIIPQGL
Rv0783c|M.tuberculosis_H37Rv QNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQSPMQSGVHIIPQGL
MUL_0501|M.ulcerans_Agy99 NNSVVTQANLTLLAFAAAYFGSSLLIPTYLQQVLHQTPMQSGLYLIPQGL
MMAR_2966|M.marinum_M TNRALTLANVAMFLYIVSTFGAGVLFPSYFQQLLDHTPLQAGMSLLPRGI
MLBr_00556|M.leprae_Br4923 HIRPFLSALGASVAAGAALMVTLVNVELFGQGVLGMDQTQAAGLLVWFLI
TH_1008|M.thermoresistible__bu RFRPFLAALGASFVAGAALMVTLVNVELFGQGVLGKDQFEAAMLLLRFLI
Mvan_1322|M.vanbaalenii_PYR-1 AESRFARSTLAAFAFMFGLGATLLAVPLYLVDRG-RSGSVAGLVLLAVPA
MAB_0284c|M.abscessus_ATCC_199 ARADIAVPLVAVFVLMLSFVAMYTSLATELVDALHLPES-MGTTIRLAGV
Mflv_2220|M.gilvum_PYR-GCK RDARFVILVLVAALGMSGLFAYIAGASFVLQGRFGLDQTAFALVFGAGAV
.
MAV_1539|M.avium_104 LLYLVGTGTSVVAGRLADRRGRPL--------------------------
MSMEG_0103|M.smegmatis_MC2_155 LAYLAGTWASARAGAEATRFGRKP--------------------------
Mb0805c|M.bovis_AF2122/97 GAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGLGTFAFGVARQADYL---
Rv0783c|M.tuberculosis_H37Rv GAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGLGTFAFGVARQADYL---
MUL_0501|M.ulcerans_Agy99 GAMLTMPIASAFMDRRGPGKSVLLGIVLIAVGLAMFTFGVATRAQYL---
MMAR_2966|M.marinum_M GAALAVPLAGALVDRRGARGVLVIGVTLIATGMGVFAFGVATQRDYL---
MLBr_00556|M.leprae_Br4923 ALPIGAVLGGWGATKAGDRTMTFVGLLITAGGYWLISHWPVDLLNYRRSI
TH_1008|M.thermoresistible__bu ALPIGALIGGWLATRVGDRIVTFVGLLIAAGGFWLISHWPVDLLAAR-HD
Mvan_1322|M.vanbaalenii_PYR-1 SMTVLGPLVGRYLDRIGPRRVLRTGLLLMAAG------------------
MAB_0284c|M.abscessus_ATCC_199 PGMLVAPFVGSWVGRVGTETGALVG-------------------------
Mflv_2220|M.gilvum_PYR-GCK ALIGTTQCNVILLKRFTPQQIMVWALAAS---------------------
MAV_1539|M.avium_104 -------VLGAALPIAVAGLLLTVPPSLAAIVAGVGVFTGGFFAAHTVAS
MSMEG_0103|M.smegmatis_MC2_155 -------VLLVSIATMIAGVAVTMSTNAVVVLVGLVIATAGFFGAHAIAS
Mb0805c|M.bovis_AF2122/97 ------PILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIARGSTLISVN
Rv0783c|M.tuberculosis_H37Rv ------PILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIARGSTLISVN
MUL_0501|M.ulcerans_Agy99 ------PTLLVGLAIMGMGTGCTMMPLAGAAVLTLKPREIARGSTLISVT
MMAR_2966|M.marinum_M ------PMLLIGLTILGMGMGCTRMPLVAVAMQSLAPNQIARGSTLIKVN
MLBr_00556|M.leprae_Br4923 FGLFSVPTMYADLLVAGLGLGLVIGPLSSATLRVVPTAQHGIASAAVVVA
TH_1008|M.thermoresistible__bu LGVVTLPVLDTDLAIVGIGLGLVIGPLTSAALRVVPAAEHGIASAAVVVA
Mvan_1322|M.vanbaalenii_PYR-1 ---------QVGLAVAVGGRIADRVHGLAGLTAILVVVGIGIAFVQTPAA
MAB_0284c|M.abscessus_ATCC_199 ----------YLLAAAGLAVEALGAGSVPIIIAGSNIFVAGIAVSSTAMV
Mflv_2220|M.gilvum_PYR-GCK ----------SVFGLVFVALAAAGVGGVFGFVIPVWAILAAMGLVIPNAP
: . .
MAV_1539|M.avium_104 GWVGAVAQRDRAEASALYLFSYYLGGSVAGAFG---------GVLYGVGG
MSMEG_0103|M.smegmatis_MC2_155 GWVGAEAGDSKAQASALYNLFYYGGSSAVGWLG---------GLAFDAAG
Mb0805c|M.bovis_AF2122/97 QQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAAVDPSS
Rv0783c|M.tuberculosis_H37Rv QQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAAVDPSS
MUL_0501|M.ulcerans_Agy99 QQVGGSIGTALMATILTNQFNRSDNIWVANKVATLEQNAAARGVTVDPSG
MMAR_2966|M.marinum_M QQMAAAVGTALMSVILTSQLNNGENASGASKAEILREHRP--GVPLDQP-
MLBr_00556|M.leprae_Br4923 RMTGMLIGVAALTAWGLYRFN--QILAGLSTAVPPDATLIERAAAVADQ-
TH_1008|M.thermoresistible__bu RMIGMVIGLAALSAWGLYRFN--QHLQSLAAREPGGDSLAARLAAEAAR-
Mvan_1322|M.vanbaalenii_PYR-1 TGATRSPAGAQGTGLGLYNLIRFSGSAMGAAWV---------AIALAAFG
MAB_0284c|M.abscessus_ATCC_199 TMFGDRAAPRRGAGTALYGFTVFLGASSAPYLAHALGFRPLLGILVAALI
Mflv_2220|M.gilvum_PYR-GCK AVALTRHPDAAGTAAALLGAVQFGGGAAIAPVVGVLGNDELAMAVVMAAG
MAV_1539|M.avium_104 -------------WSATVCFVVVLLMAGAALVALLVRDNGFRIGRRVVAG
MSMEG_0103|M.smegmatis_MC2_155 -------------WSA----VAGTVMGLAALAGLIA----FALAR-----
Mb0805c|M.bovis_AF2122/97 LPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQQAS-
Rv0783c|M.tuberculosis_H37Rv LPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQQAS-
MUL_0501|M.ulcerans_Agy99 LPPQALTPDFADNVMHDLSHAYTVVFGVVVVLVVSTLIPAASLPKTPAAA
MMAR_2966|M.marinum_M -------------VLHELSRAYSVVFVVAVVLAVLAFIPLAFLPKWSALT
MLBr_00556|M.leprae_Br4923 -------------AKRAYTMMYSDIFMITAIVCVIGALLGLLISSRKEHA
TH_1008|M.thermoresistible__bu -------------YREAYAMQYGEIFAITALLCVAGALLGLLIAGHGDRA
Mvan_1322|M.vanbaalenii_PYR-1 -----------------AATGYSAVFLVVGGVAVLGLLGSFVGPDPAPAR
MAB_0284c|M.abscessus_ATCC_199 G---------------------GAALVASSRRASGPVKPVVDISTQSSPL
Mflv_2220|M.gilvum_PYR-GCK ---------------------VIIALGALLVTGAHRNTEPARVPDEAVAE
:
MAV_1539|M.avium_104 VASVK-------------------------------------------
MSMEG_0103|M.smegmatis_MC2_155 ------------------------------------------------
Mb0805c|M.bovis_AF2122/97 ------------------HRRAPLLSA---------------------
Rv0783c|M.tuberculosis_H37Rv ------------------HRRAPLLSA---------------------
MUL_0501|M.ulcerans_Agy99 PASFEQ------------HHCDTQYGVHRV------------------
MMAR_2966|M.marinum_M ALSQTSSLERTPEPDAAPHGRQSPPEDTRPGAGRNSSSNDLTPSGPDG
MLBr_00556|M.leprae_Br4923 SEPKVPE-----------------------------------------
TH_1008|M.thermoresistible__bu DDTGTPDGSVWDLHGSGDDDTPTRVIGRQVPLPSMEQTVRMPRPPGD-
Mvan_1322|M.vanbaalenii_PYR-1 ------------------------------------------------
MAB_0284c|M.abscessus_ATCC_199 P-----------------------------------------------
Mflv_2220|M.gilvum_PYR-GCK PA----------------------------------------------