For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTTPAKATALQQNLPGRPDGSTGSSVAAGERHVDRLDGSLLRIAGICVFASVAVNLANTAVSVAQRSLIV TFGSNQAVVAWTVTAYTLTEAAAIPLSGWAADRIGTKRLFMISVLGFTLGSVLCAVAPNIACLIIFRAVQ GGGGGILMPLVITILAREAGPNRLARLMSVMGIPLLLGPMAGPILGGWLIDDYGWQWIFWINVPIGLITV ALAAIAFPGDHTAPSETLDIIGMLLLSPGLATFLYGLSTVPARGTVADRHVLIPATAGLVLMGAFVFHAL YRADRPLIDLRLFRNRVVTVANATIVFVAAGFSGAVLLVPSYFQQLLRETPLQVGIHMIPLGLGAAVTIP TSSVLMDRHGAGKVVLGGVTLISVGMGTLAFGAAEHAAYVPTLLIGLTIVGMGIGSIMLQLTTVAVQTLA PHQIARGSTLVSVNQQLSASASTALMSVILTSQFNRSANIAAANRLEALNESATRRGIPLDPSQIPPRTL NPDFGANLLHDLSQAYTVVFVYASVVMLVTFLPAVFLPMRAAITSAPLREPIDPEFPEMAARGRPG*
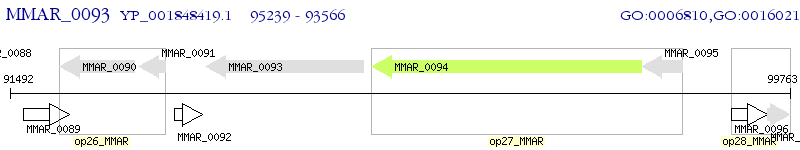
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0093 | ermB | - | 100% (557) | integral membrane efflux protein ErmB |
| M. marinum M | MMAR_2515 | - | 0.0 | 60.42% (528) | integral membrane drug efflux protein |
| M. marinum M | MMAR_4905 | - | e-173 | 58.57% (519) | integral membrane drug efflux protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 1e-169 | 58.87% (513) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_2221 | - | 5e-47 | 27.47% (495) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 1e-169 | 58.87% (513) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_01562 | - | 1e-38 | 28.87% (433) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0945 | - | 6e-46 | 28.64% (412) | EmrB/QacA family drug resistance transporter |
| M. avium 104 | MAV_0730 | - | 1e-178 | 60.12% (519) | EmrB efflux protein |
| M. smegmatis MC2 155 | MSMEG_5670 | - | 3e-43 | 30.44% (427) | efflux membrane protein |
| M. thermoresistible (build 8) | TH_2858 | - | 2e-41 | 30.19% (424) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0501 | - | 1e-171 | 57.36% (523) | integral membrane drug efflux protein |
| M. vanbaalenii PYR-1 | Mvan_4474 | - | 1e-46 | 29.85% (402) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2221|M.gilvum_PYR-GCK ---------------------------------------MFDTVTSRAAG
Mvan_4474|M.vanbaalenii_PYR-1 ---------------------------------------MFETVTARVRG
Mb0805c|M.bovis_AF2122/97 ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
Rv0783c|M.tuberculosis_H37Rv ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
MUL_0501|M.ulcerans_Agy99 ----------------------MLSTAMDEAHCTPAEVAPPDTRTGTSSP
MAV_0730|M.avium_104 MRSPSAITELAAAGAGESGLFDMLSNAMDEAPYAAAKTP-PHAPTGQPGA
MMAR_0093|M.marinum_M ----------------------MNTTPAKATALQQNLPGRPDGSTGSSVA
MAB_0945|M.abscessus_ATCC_1997 -------------------------------------------MEAAVGA
MSMEG_5670|M.smegmatis_MC2_155 -------------------------------------------MNETVGA
MLBr_01562|M.leprae_Br4923 ----------MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSK
TH_2858|M.thermoresistible__bu --------------------------------------------MEGANK
Mflv_2221|M.gilvum_PYR-GCK TTNPWP---------ALWALLIG--FFMILVDATIVAVANPAIMASLNAG
Mvan_4474|M.vanbaalenii_PYR-1 -ADPWP---------ALWALLIG--FFMILVDATIVPVANPAIMAQLGAD
Mb0805c|M.bovis_AF2122/97 GQRSYPDRLDVGLLRTAGVCVLA--SVMAHVDVTVVSVAQRTFVADFGST
Rv0783c|M.tuberculosis_H37Rv GQRSYPDRLDVGLLRTAGVCVLA--SVMAHVDVTVVSVAQRTFVADFGST
MUL_0501|M.ulcerans_Agy99 GEHVYPDRLDAGLLRIAGVCVLA--SLMSVLDATVVGIAQRTFIIEFDST
MAV_0730|M.avium_104 TEREYPDKLDAALLRISGVCILA--TVMAILDVTVVSVAQRTFIDQFSSS
MMAR_0093|M.marinum_M AGERHVDRLDGSLLRIAGICVFA--SVAVNLANTAVSVAQRSLIVTFGSN
MAB_0945|M.abscessus_ATCC_1997 LSERRK-------IVILGSCCLS--LLIVSMDSTIVNVALPTIRADFGAT
MSMEG_5670|M.smegmatis_MC2_155 LSARRK-------AIILVSCCLS--LLIVSMDATIVNVAIPSIRSDLGAS
MLBr_01562|M.leprae_Br4923 YPTLLPSGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLS
TH_2858|M.thermoresistible__bu LPTWLPSR-----RFIAAVFAIGGMQLLATMDSTIAIVALPRIQDELNLT
:. . : * . :* : :.
Mflv_2221|M.gilvum_PYR-GCK YDAVIWVTSAYLLAYAVPLLVSGRLGDRYGPKNVYMVGLTVFTVASLWCG
Mvan_4474|M.vanbaalenii_PYR-1 YDAVIWVTSAYLLAYAVPLLVSGRLGDRYGPKTVYLVGLAVFTAASLWCG
Mb0805c|M.bovis_AF2122/97 QAVVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCA
Rv0783c|M.tuberculosis_H37Rv QAVVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCA
MUL_0501|M.ulcerans_Agy99 QAVVAWTMTGYTLALATVIPVAGWAADRFGTKRLWMGSVLAFALGSLLCA
MAV_0730|M.avium_104 QAVVAWTMTGYTLALATVIPITGWAADRFGTKRLFIGSVVAFMLGSLLCA
MMAR_0093|M.marinum_M QAVVAWTVTAYTLTEAAAIPLSGWAADRIGTKRLFMISVLGFTLGSVLCA
MAB_0945|M.abscessus_ATCC_1997 TSQLQWVIDIYTLGLASLLMLSGAAGDRLGRRKVFHLGLSIFAIGSLLCS
MSMEG_5670|M.smegmatis_MC2_155 PSQLQWVIDVYTLVLASLLLLAGATGDRFGRRRTFHLGLTVFAVASLLCS
MLBr_01562|M.leprae_Br4923 DAGRSWGITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCA
TH_2858|M.thermoresistible__bu DAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVTLFTIASVLCG
* * * . : * .* * : : .: * .*: *.
Mflv_2221|M.gilvum_PYR-GCK LSDSIGMLIAARVVQGVGAALLTPQTMTVITRTFP-AHRRGVAMSVWGAT
Mvan_4474|M.vanbaalenii_PYR-1 LSDSIGMLVAARVLQGVGAALLTPQTLTVITRTFP-AHNRGVAMSVWGAT
Mb0805c|M.bovis_AF2122/97 VAPNILLLIIFRVVQGFGGGMLTPVSFAILAREAG-PKRLGRVMAVVGIP
Rv0783c|M.tuberculosis_H37Rv VAPNILLLIIFRVVQGFGGGMLTPVSFAILAREAG-PKRLGRVMAVVGIP
MUL_0501|M.ulcerans_Agy99 LAPNILSLIFFRVLQGVGGGMLMPLGFIILTRAAG-PKRLGRLMAAIGIP
MAV_0730|M.avium_104 LAANILQLIVFRVVQGIGGGMLLPLGFMILTREAG-PRRLGRLMSILSIP
MMAR_0093|M.marinum_M VAPNIACLIIFRAVQGGGGGILMPLVITILAREAG-PNRLARLMSVMGIP
MAB_0945|M.abscessus_ATCC_1997 VAPTVGLLIAARLLQAVGGSMLNPVALSIISQVFTGRVERARALGFWGAV
MSMEG_5670|M.smegmatis_MC2_155 VAPNIETLIAARLLQAIGGSMLNPVAMSIITQVFTGRVERARAIGVWGSV
MLBr_01562|M.leprae_Br4923 VAWDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAM
TH_2858|M.thermoresistible__bu IAWDEATLVIARLLQGVGAAIASPTGLALVATTFPKGPARNAATAVFAAM
:: *: * :*. *..: * : ::: . .
Mflv_2221|M.gilvum_PYR-GCK AGVATLVGPLAGGVLVDSLGWQWIFIVNVPVGIVGLVMALRLVPALPNPV
Mvan_4474|M.vanbaalenii_PYR-1 AGVATLAGPLAGGVLVDSLGWQWIFIVNLPVGVLGFALAVWLVPSLPTAR
Mb0805c|M.bovis_AF2122/97 MLLGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAA
Rv0783c|M.tuberculosis_H37Rv MLLGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAA
MUL_0501|M.ulcerans_Agy99 MLLGPIGGPILGGWLISSFGWHWIFLVNLPIGLAAFALAAITFPADRPVP
MAV_0730|M.avium_104 MLLAPIAGPILGGWLIDTSSWRWIFLINVPIGLLTVALAAVVFPRDHPAR
MMAR_0093|M.marinum_M LLLGPMAGPILGGWLIDDYGWQWIFWINVPIGLITVALAAIAFPGDHTAP
MAB_0945|M.abscessus_ATCC_1997 VGISMALGPIVGGLLIQWVGWRAVFWINLPVCVLAFVLTAVFVPETKSAT
MSMEG_5670|M.smegmatis_MC2_155 VGISMALGPLVGGALIEYVDWRAVFWINLPICALAILLTAIFVRESKSST
MLBr_01562|M.leprae_Br4923 TAIGSVMGLVVGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKER
TH_2858|M.thermoresistible__bu TGVGSVMGLVVGGALTEVS-WRLAFLVNVPIGLLVIYLARRTLRETHRER
:. * : ** * *: * :*:*: . :: .
Mflv_2221|M.gilvum_PYR-GCK RQGFDVPGVVLS--GVGLFLIVFALQEGQSHGWALWIWATVLAGVAVMAA
Mvan_4474|M.vanbaalenii_PYR-1 AQRFDLPGVVLS--GVALFLIVFALQEGQSHDWAPWIWATIGIGIAVMAA
Mb0805c|M.bovis_AF2122/97 SENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAA
Rv0783c|M.tuberculosis_H37Rv SENFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAA
MUL_0501|M.ulcerans_Agy99 SESFDVIGVLLLSPGLAAFLLALSLIPDRGTVADRYVLIPVIAGLSLIGG
MAV_0730|M.avium_104 SETFDAVGVLLLSPGLATFLFAVSSIPGRGTVADRHVLIPAAMGLTLIAG
MMAR_0093|M.marinum_M SETLDIIGMLLLSPGLATFLYGLSTVPARGTVADRHVLIPATAGLVLMGA
MAB_0945|M.abscessus_ATCC_1997 MRDIDPVGQALAVLFLFGIVFALIEGPSLGWATPGLIVVLAVALAAFAVF
MSMEG_5670|M.smegmatis_MC2_155 MRDVDPVGQGLGVAFLFGVVFVLIEGPGMGWTDPRTLAVAATALVAFLAF
MLBr_01562|M.leprae_Br4923 MK-LDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAF
TH_2858|M.thermoresistible__bu LK-LDAAGAILATLGCTAAVFAFSMGPERGWASVITIATGAAALVFLLAF
. .* * * : . . :
Mflv_2221|M.gilvum_PYR-GCK FVYWQSINTAGALIPLVIFRDRNFSMSNLGVATIGFVATGMILPLMFFAQ
Mvan_4474|M.vanbaalenii_PYR-1 FLYWQSVNTGEPLIPLIIFGDRNFSMSNLGVATIGFVATGMILPLMFYAQ
Mb0805c|M.bovis_AF2122/97 FVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQ
Rv0783c|M.tuberculosis_H37Rv FVAHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQ
MUL_0501|M.ulcerans_Agy99 FAWHAWHRADHPLIDLHLFNNSVVTQANLTLLAFAAAYFGSSLLIPTYLQ
MAV_0730|M.avium_104 FVGHAWHRADHPLIDLRLFRNPVLTHANVTMLVFATAFFGAGLLLPSYFQ
MMAR_0093|M.marinum_M FVFHALYRADRPLIDLRLFRNRVVTVANATIVFVAAGFSGAVLLVPSYFQ
MAB_0945|M.abscessus_ATCC_1997 LRYESRR--RHPFIDLRFFRSIPFASATLTAICACMAWSAFLFTMSLYLQ
MSMEG_5670|M.smegmatis_MC2_155 LRYESRR--HDPFIDLRFFRSVPFASATLVAVCAFAAYGAFLFMMSLYLQ
MLBr_01562|M.leprae_Br4923 AVVERTA--ENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQ
TH_2858|M.thermoresistible__bu LYVERTA--DNPVVPFSLFRDRNRVATFGAIFLAGGVMFTLTVLIGLYVQ
..: : :* . : . : : *
Mflv_2221|M.gilvum_PYR-GCK AVCGLTPTRAALLTAPMAVATGVLAPFVGKLVDRAHPRSVVGFGFSLMAI
Mvan_4474|M.vanbaalenii_PYR-1 SVCGLTPTQAALLTAPMAVATGVLAPLVGKLVDRSHPRPVVGFGFALMAI
Mb0805c|M.bovis_AF2122/97 QVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAA
Rv0783c|M.tuberculosis_H37Rv QVLHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAA
MUL_0501|M.ulcerans_Agy99 QVLHQTPMQSGLYLIPQGLGAMLTMPIASAFMDRRGPGKSVLLGIVLIAV
MAV_0730|M.avium_104 QVLHQTPMQAGVHMIPQGLGAMLTMRLTGPLVDRQGPGKVVLVGIALITA
MMAR_0093|M.marinum_M QLLRETPLQVGIHMIPLGLGAAVTIPTSSVLMDRHGAGKVVLGGVTLISV
MAB_0945|M.abscessus_ATCC_1997 GQRHFSAMHTGLIYLPIAVGALVFSPISGRMVGRFGARPSLLAAGTLFLT
MSMEG_5670|M.smegmatis_MC2_155 SERGYSAMFTGLIYLPIAIGALIFSPLSGRMVGRFGSRPSLVSAGVLITA
MLBr_01562|M.leprae_Br4923 DILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMV
TH_2858|M.thermoresistible__bu DIMGYSPLRAGIGFIPFVIALGIGLAASSQLVRLFPPRVLVFGGGVLVLG
:. .: * :. : . :: . . *.
Mflv_2221|M.gilvum_PYR-GCK GVTWLSIEMTPTTPIWRLLLP-LTAMGAAMAFIWSPLAATATRNLPPHLA
Mvan_4474|M.vanbaalenii_PYR-1 GLTWLSIEMTPSTPIWRLVVP-LTAMGVAMAFIWSPLAATATRNLPPQLA
Mb0805c|M.bovis_AF2122/97 GLGTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQI
Rv0783c|M.tuberculosis_H37Rv GLGTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQI
MUL_0501|M.ulcerans_Agy99 GLAMFTFGVATRAQYLPTLLVGLAIMGMGTGCTMMPLAGAAVLTLKPREI
MAV_0730|M.avium_104 GLGAFAFGVARQAPYLPTLLAGLAITGLGMGCTMMPLSVASVQALAPHQI
MMAR_0093|M.marinum_M GMGTLAFGAAEHAAYVPTLLIGLTIVGMGIGSIMLQLTTVAVQTLAPHQI
MAB_0945|M.abscessus_ATCC_1997 --ASLLLTRVSEATPIWELLVTFAIFGAGFAMVNAPITNAAVSGMPLDRA
MSMEG_5670|M.smegmatis_MC2_155 --ATVMLAQMTATTPVWLLLVIFAVFGIGFALVNAPITTAAVSGMPLDRA
MLBr_01562|M.leprae_Br4923 AMLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRI
TH_2858|M.thermoresistible__bu AMLYG-STLHADIPYFPNLVVPIVVGGIGIGVIVVPLTVSAIAGVGFDQI
: :. * . . :: : :
Mflv_2221|M.gilvum_PYR-GCK GAGSGVYNTTRQVGSVLGSAGMAALMSS-ELSTKIPGADAVPQSEGEVAA
Mvan_4474|M.vanbaalenii_PYR-1 GAGSGVYNTTRQVGSVLGSAAMAALLAS-QLSAKIPGEAPVP-VEGQAGQ
Mb0805c|M.bovis_AF2122/97 ARGSTLISVNQQVGGSIGTALMSVLLTY-QFNHSEIIATAKKVALTPESG
Rv0783c|M.tuberculosis_H37Rv ARGSTLISVNQQVGGSIGTALMSVLLTY-QFNHSEIIATAKKVALTPESG
MUL_0501|M.ulcerans_Agy99 ARGSTLISVTQQVGGSIGTALMATILTN-QFNRSDNIWVANKVATLEQNA
MAV_0730|M.avium_104 ARGTTLMSVSHQVGGSMGTALMSMILTN-QFNRSPNIVAANKLAALHQKA
MMAR_0093|M.marinum_M ARGSTLVSVNQQLSASASTALMSVILTS-QFNRSANIAAANRLEALNESA
MAB_0945|M.abscessus_ATCC_1997 GAASAVTSTSRQVGVSIGVALCGSIG---------------TPALWFMCA
MSMEG_5670|M.smegmatis_MC2_155 GAASAVASTSRQVGVSIGVALCGSVAGT-ALAGTGVDFAAASRPLWLICT
MLBr_01562|M.leprae_Br4923 GPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRA
TH_2858|M.thermoresistible__bu GPVSAIALMLQNLGGPLVLAVVQAVITSRTLYLGGTTGPVQNMDTAQLHA
. : : :.:. * :
Mflv_2221|M.gilvum_PYR-GCK LPQFLREPFALAMSQSMLLPAFVALFGVIAALFLFDLLGDRDAAAP--PA
Mvan_4474|M.vanbaalenii_PYR-1 LPAFLHAPFAAAMSQAMLLPAFVALFGVIAALFLVG-LGDRLPAPPRRPQ
Mb0805c|M.bovis_AF2122/97 AGRGAAVDPSSLPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIP
Rv0783c|M.tuberculosis_H37Rv AGRGAAVDPSSLPRQTN---FAAQLLHDLSHAYAVVFVIATALVVSTLIP
MUL_0501|M.ulcerans_Agy99 AARGVTVDPSGLPPQALTPDFADNVMHDLSHAYTVVFGVVVVLVVSTLIP
MAV_0730|M.avium_104 AAGGTPIDQSAIPRQSLAPGFGGNVLHDLSHAYTAVFVIAVALVVCTIIP
MMAR_0093|M.marinum_M TRRGIPLDPSQIPPRTLNPDFGANLLHDLSQAYTVVFVYASVVMLVTFLP
MAB_0945|M.abscessus_ATCC_1997 GLGLLIATLGIVSTSPAAVRSAQRLAPLI-EGTTAREMSRVG--------
MSMEG_5670|M.smegmatis_MC2_155 ALGVVIVAIGIVSTSPWALRSAERLAPLIGQHHRPAEVAHAR--------
MLBr_01562|M.leprae_Br4923 LDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL--
TH_2858|M.thermoresistible__bu LDQGYTYGLLWVAAVAVLVGGAALFIGYTAKQVAHAQEVKDAIDAGEL--
.
Mflv_2221|M.gilvum_PYR-GCK PDVEPLEPFDPDHPLDWADGEDDYVEYEVSREE---FDRLATDG-TADGA
Mvan_4474|M.vanbaalenii_PYR-1 ETAEPVPAFDPDDELYWADGEDEYVEYEVPWDDGAEPRRPAQEADTVDAA
Mb0805c|M.bovis_AF2122/97 AAFLPKQQAS-------HRRAPLLSA------------------------
Rv0783c|M.tuberculosis_H37Rv AAFLPKQQAS-------HRRAPLLSA------------------------
MUL_0501|M.ulcerans_Agy99 AASLPKTPAAAPASFEQHHCDTQYGVHRV---------------------
MAV_0730|M.avium_104 ASFLPKKPAA--------ETAGK---------------------------
MMAR_0093|M.marinum_M AVFLPMRAAITSAPLR-EPIDPEFPEMAARGRPG----------------
MAB_0945|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5670|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
TH_2858|M.thermoresistible__bu --------------------------------------------------
Mflv_2221|M.gilvum_PYR-GCK AYN-ITEDFTEDITD-----VLDTVVEAPPAPADSTPSQQWRDLVDQLLD
Mvan_4474|M.vanbaalenii_PYR-1 ADTDVLDPVTEPMHDGGGYPAPESAPRRDHADPTSHPDREWRSILDQLLD
Mb0805c|M.bovis_AF2122/97 --------------------------------------------------
Rv0783c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0501|M.ulcerans_Agy99 --------------------------------------------------
MAV_0730|M.avium_104 --------------------------------------------------
MMAR_0093|M.marinum_M --------------------------------------------------
MAB_0945|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5670|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
TH_2858|M.thermoresistible__bu --------------------------------------------------
Mflv_2221|M.gilvum_PYR-GCK VSSPPGAP----RNGVGSADAEHPGGGPRV-----
Mvan_4474|M.vanbaalenii_PYR-1 PPKPSDDPTGQGRNGFHVTPSEDGRGASRGRHSLD
Mb0805c|M.bovis_AF2122/97 -----------------------------------
Rv0783c|M.tuberculosis_H37Rv -----------------------------------
MUL_0501|M.ulcerans_Agy99 -----------------------------------
MAV_0730|M.avium_104 -----------------------------------
MMAR_0093|M.marinum_M -----------------------------------
MAB_0945|M.abscessus_ATCC_1997 -----------------------------------
MSMEG_5670|M.smegmatis_MC2_155 -----------------------------------
MLBr_01562|M.leprae_Br4923 -----------------------------------
TH_2858|M.thermoresistible__bu -----------------------------------