For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNYSRARGRRAGKPDTRAKILEVARRRFLEGGYQGVKLRSVAAEAGVDLALISYYFGSKRGLVGEALALS ANPADVLDQAVAKGDPATFPQRVLAGLLALWEDPASGASLRALVAGAAHDPALANRVKEMVERELIDKIA AQLSGTDARKRASMFCSQIAGLIVTRYILCLEPACSMTHDEIIRQYAPLLHLALRSTTATARRPAPNTLS GNTTGR
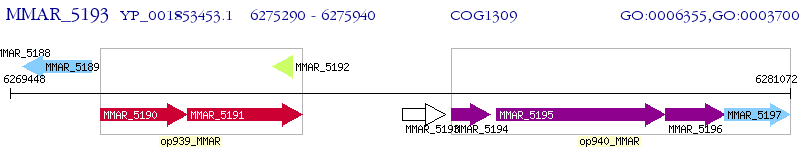
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5193 | - | - | 100% (216) | hypothetical protein MMAR_5193 |
| M. marinum M | MMAR_3397 | - | 1e-25 | 34.76% (187) | putative regulatory protein |
| M. marinum M | MMAR_4115 | - | 3e-23 | 36.61% (183) | putative regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1711c | - | 3e-20 | 32.29% (192) | hypothetical protein Mb1711c |
| M. gilvum PYR-GCK | Mflv_0253 | - | 5e-26 | 33.51% (185) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1685c | - | 4e-20 | 32.29% (192) | hypothetical protein Rv1685c |
| M. leprae Br4923 | MLBr_00717 | - | 3e-05 | 29.77% (131) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2351c | - | 3e-16 | 32.09% (187) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3088 | - | 3e-16 | 31.77% (192) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5282 | - | 4e-23 | 37.23% (188) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3542 | - | 9e-21 | 29.48% (173) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_3981 | - | 3e-23 | 36.61% (183) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0641 | - | 2e-25 | 33.17% (202) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5282|M.smegmatis_MC2_155 -MVKRSARATPGPRDDRGVLAARILAAARETFAESGWAGTTIRAVARAAD
MUL_3981|M.ulcerans_Agy99 -MGGR--RVKPGPRDEKGVLAAQILSVARDSFAENGSAGTTVRSVARAAD
Mb1711c|M.bovis_AF2122/97 MAAPDNSRRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAAKAG
Rv1685c|M.tuberculosis_H37Rv MAAPDNSRRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAAKAG
MAV_3088|M.avium_104 MTTTGTTRRRRGRPAGSSGSRERILASARELFARNGIRNTSIRAVAAAAG
MAB_2351c|M.abscessus_ATCC_199 MSS-----KRPGRRPGTSSARDDILASARKLFSLNGIDKTSIRAIASDAG
Mflv_0253|M.gilvum_PYR-GCK MPPPAQTGRRRGRRKGDPVSRETVLAAAKGRFARDGFEKTTLRAIAEDAG
Mvan_0641|M.vanbaalenii_PYR-1 MPRPGETARRRGRRQGEPVSKEAVLAAAKTRFARDGYEKTTLRAIADDAR
TH_3542|M.thermoresistible__bu -----------------------VLAAAKRRFAQDGYEKTTLRAIARDAH
MMAR_5193|M.marinum_M ----MNYSRARGRRAGKPDTRAKILEVARRRFLEGGYQGVKLRSVAAEAG
MLBr_00717|M.leprae_Br4923 -------MTTARRRLSPEDRRAELLALGAEVFGKRPYDEVRIDEIAERAG
:* . * . : :* *
MSMEG_5282|M.smegmatis_MC2_155 VDPALVYHYYGSKEGLLDAATDPPQKWLDAVVATWQAP--VGDTGRAVLR
MUL_3981|M.ulcerans_Agy99 VDPSLIYHYFGSKEGLLDAATMPPQRWLDTVAETWTTP--KADLGRRLLQ
Mb1711c|M.bovis_AF2122/97 VDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREAP--VEELGYKLPS
Rv1685c|M.tuberculosis_H37Rv VDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREAP--VEELGYKLPS
MAV_3088|M.avium_104 VDSALVHHYFGTKEKLFAAAVQIPIDPMQVIGPLREVP--VDDLGYALPS
MAB_2351c|M.abscessus_ATCC_199 VDPALVHHYFGTKLDLFREVVQLPVDPSVVLQPLRDVP--VDELGVTIPR
Mflv_0253|M.gilvum_PYR-GCK VDASMVLYLFGSKEALFRESLRLILDPGVLIEALSGGE---ADIGTRMVR
Mvan_0641|M.vanbaalenii_PYR-1 VDASMVLYLFGSKEQLFREALRLILDPEVLVAALAGVEGDSSDVGTRMVR
TH_3542|M.thermoresistible__bu VDASMVLYLFGSKAELFRESLELILDPEMLVAAMTGDD---GDVGTRMVR
MMAR_5193|M.marinum_M VDLALISYYFGSKRGLVGEALALSANPADVLDQAVAKG-DPATFPQRVLA
MLBr_00717|M.leprae_Br4923 VSRALMYHYFPDKRAFFAAVVKDEADRLYAATNIPPPAGLTMFEEVRVGV
*. ::: : : * :. :
MSMEG_5282|M.smegmatis_MC2_155 LLLDSWADDEVGPVLRAILQTAAHEP---TTHEKLRKVVERSLIGVIATM
MUL_3981|M.ulcerans_Agy99 TLLDSWADDEIGPSLRAILLTAAHED---ATRLKLRRIVESSLIG-VSQL
Mb1711c|M.bovis_AF2122/97 LLLPIWDS-ELGAGLIATLRSLISG----SDVGLARSFLEEVVTVELGSR
Rv1685c|M.tuberculosis_H37Rv LLLPIWDS-ELGAGLIATLRSLISG----SDVGLARSFLEEVVTVELGSR
MAV_3088|M.avium_104 MLLPLWDS-EVGAAFIATLRSILAG----EEISLFRTFIQDVIGVEVGPR
MAB_2351c|M.abscessus_ATCC_199 LIIALWDS-ELGANMLAVFRSALTG----ADDGLVRVFFREVLVNIIAER
Mflv_0253|M.gilvum_PYR-GCK AYLGVWESPESAESMRTMLASATSNP---DAREAFRGFIQDYVLAAVVGT
Mvan_0641|M.vanbaalenii_PYR-1 TYLGIWEAADSAASMRAMLASATSNP---DAHEAFRSFMQDYVLTAVSGV
TH_3542|M.thermoresistible__bu VYLQIWESPDTGPTIRAMLQSATSNT---DAREAFRSFLREYLLTAVSGV
MMAR_5193|M.marinum_M GLLALWEDPASGASLRALVAGAAHDP---ALANRVKEMVERELIDKIAAQ
MLBr_00717|M.leprae_Br4923 LAYMAYHKQNPEAAWAAYVGLGRSDPVLLGIDDEAKNRQMEHIMSRIGDV
: : . : : :
MSMEG_5282|M.smegmatis_MC2_155 GDD-ERVRRTRAGLVASQTIGFALMRYVWRIEPVASMREDEVLDAVGPTL
MUL_3981|M.ulcerans_Agy99 DTD-EDDRLTRSGLIASQLIGLAMTRYVWQIEPIASMSDSEVLAAITPNL
Mb1711c|M.bovis_AF2122/97 VDNPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNL
Rv1685c|M.tuberculosis_H37Rv VDNPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNL
MAV_3088|M.avium_104 VDNPAGSGVIRIQFVASQLVGVVMARYILELEPFASLPPEQIAATIAPNL
MAB_2351c|M.abscessus_ATCC_199 VDSPPGSGVLRAEFAITQMAGILVGRYIMAIEPLASLTAEQIALTVGPNI
Mflv_0253|M.gilvum_PYR-GCK LGG-DDRARLRAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTV
Mvan_0641|M.vanbaalenii_PYR-1 LGG-GEQARLRAMLAASSLVGTAMLRYVMRVAPLAELPTEDVVRLVAPTV
TH_3542|M.thermoresistible__bu LGG-GEQARLRAELAATGLVGTAMLRYIMRVPPLAELSTEEVVRMLAPTV
MMAR_5193|M.marinum_M LSG--TDARKRASMFCSQIAGLIVTRYILCLEPACSMTHDEIIRQYAPLL
MLBr_00717|M.leprae_Br4923 VSVVPESKLHPDIERDLRVIVHGWLAFTFEVCRQRIMDPSTDADRLADSC
: : : .
MSMEG_5282|M.smegmatis_MC2_155 QRYIDGDIG----AAGQIRTGFDTA-----
MUL_3981|M.ulcerans_Agy99 QRYIDGDLHPTKNSPAQRPRRTRRP-----
Mb1711c|M.bovis_AF2122/97 QRYLTGELPDDLAP----------------
Rv1685c|M.tuberculosis_H37Rv QRYLTGELPDDLAP----------------
MAV_3088|M.avium_104 QRYLTGDLPDGLAP----------------
MAB_2351c|M.abscessus_ATCC_199 QRYLTGALPS-ITP----------------
Mflv_0253|M.gilvum_PYR-GCK TRYLTAPADELGLPG---------------
Mvan_0641|M.vanbaalenii_PYR-1 TRYLTADARELGLPD---------------
TH_3542|M.thermoresistible__bu NRYLTADADELGLPSSLTAGS---------
MMAR_5193|M.marinum_M HLALRSTTATARRPAPNTLSGNTTGR----
MLBr_00717|M.leprae_Br4923 AHALLDAIARVPEISKELADAMAIARQPPQ
: