For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADHRDQAGTDDSGTRRTEILQTAAALIASSGLRTSLQEIADAAGILPGSLYHHFESKEAILIELIRRYQ DDLHQIGQSWQAKLDQPDSRTVAEKITQLGAAIANCAVAHRAALQMSFYEGPSADPELMKLTSQRPLAIQ EAMLQTLRAGRWSGCIRTEIDLPTLADRICQTMLQVGLDMMRRNASADQVSGLMCRTILQGLASHAPTDV DLDGSNALSAANDVIATWIDEKDADPSDKAAHVRAVARIEFGRKGYEVTTIRDIASASGLATGTVYRVIG SKDKLLASIMRSFGQKVEAGWVAVLRSNATPIEKLDALSWVNINALDQFSDEFRIQLAWMRQSPPDTPNP GWLYAARLRQMKSLLSEGLRSAEIQIEPPSITMLARCVIGLQWIPENILHFVGTRAALIHARDTVLRGVA VRGQ
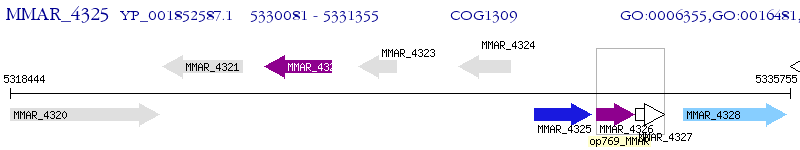
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4325 | - | - | 100% (424) | TetR family transcriptional regulator |
| M. marinum M | MMAR_5046 | - | 2e-11 | 52.38% (63) | TetR family transcriptional regulator |
| M. marinum M | MMAR_2873 | mce3R | 2e-08 | 22.70% (304) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3587c | - | 8e-12 | 56.90% (58) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4158 | - | 1e-156 | 65.88% (422) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3557c | - | 9e-12 | 56.90% (58) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00316 | - | 3e-05 | 38.18% (55) | putative TetR/AcrR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0599 | - | 5e-14 | 33.33% (165) | TetR family transcriptional regulator |
| M. avium 104 | MAV_1265 | - | 0.0 | 79.47% (414) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6009 | - | 2e-12 | 35.53% (152) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3536 | - | 1e-162 | 69.34% (411) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0135 | - | 0.0 | 97.37% (342) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0719 | - | 1e-12 | 25.40% (315) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3587c|M.bovis_AF2122/97 ---------------MDRVAGQVN-------------SRRGELLELAAAM
Rv3557c|M.tuberculosis_H37Rv ---------------MDRVAGQVN-------------SRRGELLELAAAM
MSMEG_6009|M.smegmatis_MC2_155 --------------MAPDTPSQPA-------------SRRDELLQLAATM
MAB_0599|M.abscessus_ATCC_1997 ----MDKVSEVRESDALRAGGSPQP------------SRRDELLSLAAEM
MMAR_4325|M.marinum_M ------------MADHRDQAGTD-----------DSGTRRTEILQTAAAL
MUL_0135|M.ulcerans_Agy99 ------------MADHRDQAGTD-----------DSGTRRTEILQTAAAL
MAV_1265|M.avium_104 ----------MAKAESLRTGSTA-----------DSPTRRAEILATAASL
TH_3536|M.thermoresistible__bu -----VVGRPQLTSEQRNVRKSAHTLASVAGRAYDDGTRRTEILQTAARL
Mflv_4158|M.gilvum_PYR-GCK MAADAAGQKPGRGRRAGRKRSGSAELAAVRDAAPADTTRRAEILKTANTV
Mvan_0719|M.vanbaalenii_PYR-1 ---------MASTADDHGIRRRPK-------------DRKAQIARASAEA
MLBr_00316|M.leprae_Br4923 ----MSVPRSGLSVQMAQPARPLR---------ADAARNRARILGVAYEA
.: .: :
Mb3587c|M.bovis_AF2122/97 FAERGLRATTVRDIADGAGILSGSLYHHFASKEEMVDELLRGFLDWLFAR
Rv3557c|M.tuberculosis_H37Rv FAERGLRATTVRDIADGAGILSGSLYHHFASKEEMVDELLRGFLDWLFAR
MSMEG_6009|M.smegmatis_MC2_155 FADRGLKATTVRDIADSAGILSGSLYHHFKSKEQMVEEVLRDFLDWLFGR
MAB_0599|M.abscessus_ATCC_1997 FAERGLRATTVRDIADAAGILSGSLYHHFDSKEAIVDELLRDFLEGLFAR
MMAR_4325|M.marinum_M IASSGLR-TSLQEIADAAGILPGSLYHHFESKEAILIELIRRYQDDLHQI
MUL_0135|M.ulcerans_Agy99 IASSGLR-TSLQEIADAAGILPGSLYHHFESKEAILIELIRRYQDDLHQI
MAV_1265|M.avium_104 IASSGLR-TSLQEIADAAGILPGSLYHHFESKEAILVELIRRYQDDLERI
TH_3536|M.thermoresistible__bu IASSGLR-TSLQEIADAAGILPGSLYHHFESKEAILVELLRRYHTDLDTI
Mflv_4158|M.gilvum_PYR-GCK IAATGLR-SSLQQIADAAGILAGSLYHHFDSKEAILVELIRLYHDDLDRI
Mvan_0719|M.vanbaalenii_PYR-1 FSALGYYGVSMEMIASRVGISAAALYRHYSSKYELFRDAVLNLGQQLVDG
MLBr_00316|M.leprae_Br4923 FATEGLS-VPIDEIARRAGVGAGTVYRHFPTKEALCAAVIGDRMPHLVDD
:: * .: ** .*: ..::*:*: :* : : *
Mb3587c|M.bovis_AF2122/97 YRDIVDSTANP-----LERLQGLFMASFEAIEHHHAQVVIYQDEAQRLAS
Rv3557c|M.tuberculosis_H37Rv YRDIVDSTANP-----LERLQGLFMASFEAIEHHHAQVVIYQDEAQRLAS
MSMEG_6009|M.smegmatis_MC2_155 YQQILDTATSP-----LEKLTGLFMASFEAIEHRHAQVVIYQDEAKRLSD
MAB_0599|M.abscessus_ATCC_1997 YREIAAANLNP-----VDTLKGLFLASFDAIETKHAQVAIYQAEAPRLLS
MMAR_4325|M.marinum_M GQSWQAKLDQPDSRTVAEKITQLGAAIANCAVAHRAALQMSFYEGP-SAD
MUL_0135|M.ulcerans_Agy99 GQSWQAKLDQPDSRTVAEKITQLGAAIANCAVAHRAALQMSFYEGP-SAD
MAV_1265|M.avium_104 GRTAQARLDEPDSRPVPEQIIELGSAIANCAVEHRAALQMSFYEGP-GTD
TH_3536|M.thermoresistible__bu AAHAQERLDDPDPRSPSDRITDLGVAIARCAVTHRAALQMTFYEGP-TSH
Mflv_4158|M.gilvum_PYR-GCK GDLALRKLDEPDPEPVSAQITALGCAIARCAVEHRAALQMSFYEGP-SAD
Mvan_0719|M.vanbaalenii_PYR-1 TAGIEG--DDP-----QVVLDRLVSALTDTALANRGSGGLYRWEGRYLRG
MLBr_00316|M.leprae_Br4923 GYVLLKSAGPG-----EALFTYLRSLVLHWGATDRGLVDALAGAGG----
: * :. .
Mb3587c|M.bovis_AF2122/97 QPRFSYIEDRNKQQRKMWVDVLNQGIEEGYFRPDLDVDLVYRFIRDTTWV
Rv3557c|M.tuberculosis_H37Rv QPRFSYIEDRNKQQRKMWVDVLNQGIEEGYFRPDLDVDLVYRFIRDTTWV
MSMEG_6009|M.smegmatis_MC2_155 LPQFDFVETRNKEQRKMWVDILQEGVADGSFRPDLDVDLVYRFIRDTTWV
MAB_0599|M.abscessus_ATCC_1997 QERFAYLNELNTEQRQLWLEVLRDGIAQGYFRPDLDVAVVYRFIRDATWV
MMAR_4325|M.marinum_M PELMKLTSQRPLAIQEAMLQTLRAGRWSGCIRTEIDLPTLADRICQT-ML
MUL_0135|M.ulcerans_Agy99 PELMKLTSQRPLAIQEAMLQTLRAGRWSGYIRTEIDLPTFADRICQT-ML
MAV_1265|M.avium_104 PELTKLTRQRPVAIQEAMLQTLRAGRWSGYIKPDIDLPTLADRICQT-ML
TH_3536|M.thermoresistible__bu PEVTALAQQRPTGILEAMLQTLRAARWSGYLRADIDLPILADRIVQT-ML
Mflv_4158|M.gilvum_PYR-GCK PELQALSARAPEKVQAAMLQALRAGRWSGEIRSDVDLPTLADRLCQS-ML
Mvan_0719|M.vanbaalenii_PYR-1 DDQATLLQQIHTVHH-------RIHRPLSALRPELTARQRSTLSTAALSI
MLBr_00316|M.leprae_Br4923 IDILGVAPDAEDAFLTILSDLLYAAQYAGTARTDVGVREVKSILVGC---
. :.::
Mb3587c|M.bovis_AF2122/97 SVRWYRPGGPLTAQQVGQQYLAIVLGGITKEGV-----------------
Rv3557c|M.tuberculosis_H37Rv SVRWYRPGGPLTAQQVGQQYLAIVLGGITKEGV-----------------
MSMEG_6009|M.smegmatis_MC2_155 SVRWYKPGGPLSAEQVGQQYLAIVLGGITQSQGDKHA-------------
MAB_0599|M.abscessus_ATCC_1997 SVRWFRPGGSRTAQEVAEQYLSIVLGGITVGSFSNSPEK-----------
MMAR_4325|M.marinum_M QVGLDMMRRNASADQVSGLMCRTILQGLASHAPTDVDLDGSNALSAANDV
MUL_0135|M.ulcerans_Agy99 QVGLDMMRRNASADQVSGLMCRTILQALASHAPTDVDPDGSNALSAANDV
MAV_1265|M.avium_104 QVGLDVMRHTASADQVAELLCRIILQGLASRPPSDAALDRSKAFVAANEV
TH_3536|M.thermoresistible__bu QVGLDVIRHNASPDKAATLLCRILLEGLATVLPTDEELDGSAAFRAADEV
Mflv_4158|M.gilvum_PYR-GCK HVGLDVIRHKARTDDLATLKCRIALNGLASEAPSDASLDRSKAFAAAQSV
Mvan_0719|M.vanbaalenii_PYR-1 VGSIVDHRAKLPAAQVHALIAQISRAVLGAELSADPPRYPPDVVPAR---
MLBr_00316|M.leprae_Br4923 ------QAMEAYNSALAERVTDVVVDGLRAAR------------------
:
Mb3587c|M.bovis_AF2122/97 --------------------------------------------------
Rv3557c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6009|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0599|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4325|M.marinum_M IATWIDEKDADPSDKAAHVRAVARIEFGRKGYEVTTIRDIASASGLATGT
MUL_0135|M.ulcerans_Agy99 IATWIDEKDTDPSDKAAHVRAVARMEFGRKGYEVTTVRDIASASGLGTGT
MAV_1265|M.avium_104 IETWSDDSDADPGDKAALVRAVARAEFGRRGYEVTTIRDIAAAAGLGTGT
TH_3536|M.thermoresistible__bu VQSWSLEAEAEADDKAAHVRAVARAEFGRRGYEGTTIRDIARAAGLGTGT
Mflv_4158|M.gilvum_PYR-GCK ISTWADD-EDETDLKAAHLRAVARAEFGRRGYEMTTIRDIASAAGLGTGT
Mvan_0719|M.vanbaalenii_PYR-1 --------PTAEPSKYEALLTASMQLFDRNGYRDTTMEDIASAVGMPTSG
MLBr_00316|M.leprae_Br4923 --------------------------------------------------
Mb3587c|M.bovis_AF2122/97 --------------------------------------------------
Rv3557c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6009|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0599|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4325|M.marinum_M VYRVIGSKDKLLASIMRSFGQKVEAGWVAVLRSNATPIEKLDALSWVNIN
MUL_0135|M.ulcerans_Agy99 VYRVIGSKDKLLASIMRSFGQKVEAGWVAVRRSNATPIEKLDALSWVNIN
MAV_1265|M.avium_104 VYRVIGSKDELLDSIMRSFGKKVEAGWVSVLRSDATPTEKLDALSWVNVN
TH_3536|M.thermoresistible__bu VYRLIGSKEELLASIMRSFGEKIAVGWTDVLRTDAPAIAKLDALSWVNTN
Mflv_4158|M.gilvum_PYR-GCK VYRIIGSKDELLMSIMLEFGRKVGGAWSDIVRTDSTTVEKLDALSWLNIN
Mvan_0719|M.vanbaalenii_PYR-1 IYKYFSGKSDILATIFRRAADRVSAELASVVATMSDPEEALTAVIDGYVQ
MLBr_00316|M.leprae_Br4923 --------------------------------------------------
Mb3587c|M.bovis_AF2122/97 --------------------------------------------------
Rv3557c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6009|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0599|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4325|M.marinum_M -ALDQFSDEFRIQLAWMRQSPPDTPNPGWLYAARLRQMKSLLSEGLRSAE
MUL_0135|M.ulcerans_Agy99 -ALDQFSDEFRIQLAWMRQ-------------------------------
MAV_1265|M.avium_104 -ALDQFSDEFRIQLAWMRLSPP-TANPGWSYATRLRQMKSLLSEGLRTGE
TH_3536|M.thermoresistible__bu -ALDRFGDEFRIQLAWMRQSPPDTPNPGWLFAKRVRQVKSLLAAAIRSGE
Mflv_4158|M.gilvum_PYR-GCK -ALDQFPDEFRIQLAWLRQSPPDSANPAWSFTTRIRQMKSLLSEGIRAGE
Mvan_0719|M.vanbaalenii_PYR-1 RSFDQPELECVYYSERLNMSAADQRIVRDLQRSTVESWVELVVAVRPEWS
MLBr_00316|M.leprae_Br4923 --------------------------------------------------
Mb3587c|M.bovis_AF2122/97 --------------------------------------------------
Rv3557c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6009|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0599|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4325|M.marinum_M IQIEPPSITMLARCVIGLQWIPENILHFVGTRAALIHARDTVLRGVAVRG
MUL_0135|M.ulcerans_Agy99 --------------------------------------------------
MAV_1265|M.avium_104 IAIAAPSTAMLARCLIALQWIPENILAEIGKRPALLHVRDTVLRGVAVRG
TH_3536|M.thermoresistible__bu ISIDSPSNELLARSVIGVGWVPENILRQIGTRAAQILVRDTVLRGAADRD
Mflv_4158|M.gilvum_PYR-GCK IHIDSPTADMLARCIIGEQWIPENILQEVGPRNALILARDTILRGVAVRT
Mvan_0719|M.vanbaalenii_PYR-1 AAEARFAVHAAMALVVDLGRLMRYENSEEARETVSGLVDMTLLGRYRLRA
MLBr_00316|M.leprae_Br4923 --------------------------------------------------
Mb3587c|M.bovis_AF2122/97 -----
Rv3557c|M.tuberculosis_H37Rv -----
MSMEG_6009|M.smegmatis_MC2_155 -----
MAB_0599|M.abscessus_ATCC_1997 -----
MMAR_4325|M.marinum_M Q----
MUL_0135|M.ulcerans_Agy99 -----
MAV_1265|M.avium_104 AAVDN
TH_3536|M.thermoresistible__bu VTR--
Mflv_4158|M.gilvum_PYR-GCK -----
Mvan_0719|M.vanbaalenii_PYR-1 TLPAK
MLBr_00316|M.leprae_Br4923 -----