For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRRVNTVAMGSSPSSSSTRLSSPAGDPHTGLRAVTECTGSAVVVHVGGDVDASNEVVWQRLVNRSAAIAI APGPFVIDIRDLDFIGSCAYAVLAQESVRCRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALS PVPNGH
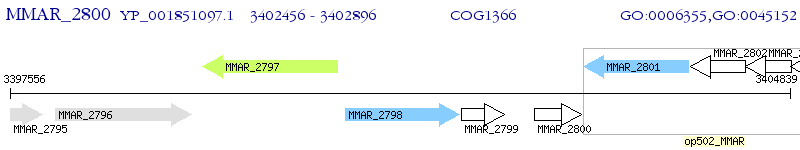
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2800 | - | - | 100% (146) | hypothetical protein MMAR_2800 |
| M. marinum M | MMAR_2061 | - | 2e-31 | 50.00% (130) | hypothetical protein MMAR_2061 |
| M. marinum M | MMAR_5181 | - | 4e-06 | 25.22% (115) | anti-anti-sigma regulatory factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1939 | - | 2e-62 | 79.72% (143) | hypothetical protein Mb1939 |
| M. gilvum PYR-GCK | Mflv_3879 | - | 7e-09 | 31.01% (129) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv1904 | - | 1e-62 | 79.72% (143) | hypothetical protein Rv1904 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3518 | - | 1e-35 | 50.00% (132) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | MSMEG_1786 | - | 0.0001 | 29.77% (131) | STAS domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2925 | - | 6e-78 | 100.00% (143) | hypothetical protein MUL_2925 |
| M. vanbaalenii PYR-1 | Mvan_2530 | - | 2e-08 | 32.41% (108) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1939|M.bovis_AF2122/97 ---MRTVAIGPGAGPSSTRPSSQPSDL-HSGLRAVTECTGSAVVVHVGGD
Rv1904|M.tuberculosis_H37Rv ---MRTVAIGPGAGPSSTRPSSQPSDL-HSGLRAVTECTGSAVVVHVGGD
MMAR_2800|M.marinum_M MRRVNTVAMGSSPSSSSTRLSSPAGDP-HTGLRAVTECTGSAVVVHVGGD
MUL_2925|M.ulcerans_Agy99 ---MNTVAMGSSPSSSSTRLSSPAGDP-HTGLRAVTECTGSAVVVHVGGD
MAV_3518|M.avium_104 -----MEPMSATHLTLSTKLVYELGDP-NSTLRATTARSGSAVLIYAGGE
Mflv_3879|M.gilvum_PYR-GCK -----MVAIRTERIFAPSISSDEDQTRCGAAIFATRTCSPNRIAVAVVGE
Mvan_2530|M.vanbaalenii_PYR-1 -----MVAVGTKTIYRPLTAVTDEQERCGHATFAVRRCSAHRMAIAVIGE
MSMEG_1786|M.smegmatis_MC2_155 -----MPTISVAKRSSPHSGSHAPQR----AHFTTEQINTATAVVTVHGD
.: . :. . : . *:
Mb1939|M.bovis_AF2122/97 IDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESVR
Rv1904|M.tuberculosis_H37Rv IDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESVR
MMAR_2800|M.marinum_M VDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESVR
MUL_2925|M.ulcerans_Agy99 VDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESVR
MAV_3518|M.avium_104 IDACNEDTWRHLVSEAAGVVATPGPFVVDVTGLEFMGCCAFAVLTDEAKR
Mflv_3879|M.gilvum_PYR-GCK VDAVNGRELARYVERHLRVSRQ---LVLDLRAVDFFGSQGFTALYYISVH
Mvan_2530|M.vanbaalenii_PYR-1 IDAVNGRALGRYVERHTRISRQ---LVLDLRAVDFFGSQGFTALYYISVH
MSMEG_1786|M.smegmatis_MC2_155 LDASNAGLLTDYALKALTRNAK---LVLDLSDVAFFGAACFTALHTLNVR
:** * . . :*:*: : *:*. ::.* :
Mb1939|M.bovis_AF2122/97 CRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYAMVETALAPPPSAH---
Rv1904|M.tuberculosis_H37Rv CRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYATVETALAPPPSAH---
MMAR_2800|M.marinum_M CRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH---
MUL_2925|M.ulcerans_Agy99 CRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH---
MAV_3518|M.avium_104 CRQRGIDLRLVSCEPIVGRIIDACGLSDILPIYPTVDSALSGADRW----
Mflv_3879|M.gilvum_PYR-GCK CTRSDVDWAIVAS-PPVRRLLSICDPESELPLADDLTAALARLDRRAQRH
Mvan_2530|M.vanbaalenii_PYR-1 CARSDVDWVIVGS-PAVRRLLAICDPDGELPLTDALSSALARLDRLAHGG
MSMEG_1786|M.smegmatis_MC2_155 CAGADAEWVVVPS-KAVTRVLRICDPDSGLTTAPDVVSALSRKS--DQRP
* . : :* * * : *. :. : :**:
Mb1939|M.bovis_AF2122/97 --------------
Rv1904|M.tuberculosis_H37Rv --------------
MMAR_2800|M.marinum_M --------------
MUL_2925|M.ulcerans_Agy99 --------------
MAV_3518|M.avium_104 --------------
Mflv_3879|M.gilvum_PYR-GCK TPFVWSARSVWRTG
Mvan_2530|M.vanbaalenii_PYR-1 QHLVWTAKSGWHAR
MSMEG_1786|M.smegmatis_MC2_155 LQLVENT-------