For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTVAIGPGAGPSSTRPSSQPSDLHSGLRAVTECTGSAVVVHVGGDIDASNEVAWQRLVSKSAAIAIAPG PFVIDIRDLDFMGSCAYAVLAQESVRCRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYATVETALAPPP SAH
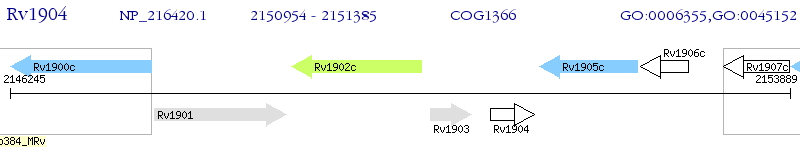
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1904 | - | - | 100% (143) | hypothetical protein Rv1904 |
| M. tuberculosis H37Rv | Rv2638 | - | 2e-34 | 52.80% (125) | hypothetical protein Rv2638 |
| M. tuberculosis H37Rv | Rv1365c | rsfA | 6e-29 | 48.78% (123) | anti-anti-sigma factor RSFA (anti-sigma factor antagonist) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1939 | - | 9e-78 | 99.30% (143) | hypothetical protein Mb1939 |
| M. gilvum PYR-GCK | Mflv_4938 | - | 3e-08 | 29.25% (106) | anti-sigma-factor antagonist |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2800 | - | 2e-62 | 79.72% (143) | hypothetical protein MMAR_2800 |
| M. avium 104 | MAV_3518 | - | 6e-34 | 51.61% (124) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0498 | - | 5e-05 | 31.08% (74) | anti-anti-sigma factor |
| M. ulcerans Agy99 | MUL_2925 | - | 1e-62 | 79.72% (143) | hypothetical protein MUL_2925 |
| M. vanbaalenii PYR-1 | Mvan_2530 | - | 1e-08 | 34.26% (108) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1904|M.tuberculosis_H37Rv ---MRTVAIGPGAGPSSTRPSSQPSDLH-SGLRAVTECTGSAVVVHVGGD
Mb1939|M.bovis_AF2122/97 ---MRTVAIGPGAGPSSTRPSSQPSDLH-SGLRAVTECTGSAVVVHVGGD
MMAR_2800|M.marinum_M MRRVNTVAMGSSPSSSSTRLSSPAGDPH-TGLRAVTECTGSAVVVHVGGD
MUL_2925|M.ulcerans_Agy99 ---MNTVAMGSSPSSSSTRLSSPAGDPH-TGLRAVTECTGSAVVVHVGGD
MAV_3518|M.avium_104 -----MEPMSATHLTLSTKLVYELGDPN-STLRATTARSGSAVLIYAGGE
Mflv_4938|M.gilvum_PYR-GCK -----------------MMMISTDQTRS-GKFTVTPRRDGDLLVLHVTGD
TH_0498|M.thermoresistible__bu --------------------------------VIDERWDADIAVVSATGV
Mvan_2530|M.vanbaalenii_PYR-1 -----MVAVGTKTIYRPLTAVTDEQERCGHATFAVRRCSAHRMAIAVIGE
. : . *
Rv1904|M.tuberculosis_H37Rv IDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESVR
Mb1939|M.bovis_AF2122/97 IDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESVR
MMAR_2800|M.marinum_M VDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESVR
MUL_2925|M.ulcerans_Agy99 VDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESVR
MAV_3518|M.avium_104 IDACNEDTWRHLVSEAAGVVATPGPFVVDVTGLEFMGCCAFAVLTDEAKR
Mflv_4938|M.gilvum_PYR-GCK LDVLTAPTLSTHLD--IALADAPPVVIVDISDVSFLSSAGIGLLVETHRL
TH_0498|M.thermoresistible__bu VDMLSAPQLDEAIR--TALGKKPRALVVDFTAVDFLASAGMGVLVAIHDE
Mvan_2530|M.vanbaalenii_PYR-1 IDAVNGRALGRYVERHTRIS---RQLVLDLRAVDFFGSQGFTALYYISVH
:* . : : .::*. :.*:.. . *
Rv1904|M.tuberculosis_H37Rv CRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYATVETALAPPPSAH---
Mb1939|M.bovis_AF2122/97 CRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYAMVETALAPPPSAH---
MMAR_2800|M.marinum_M CRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH---
MUL_2925|M.ulcerans_Agy99 CRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH---
MAV_3518|M.avium_104 CRQRGIDLRLVSCEPIVGRIIDACGLSDILPIYPTVDSALSGADRW----
Mflv_4938|M.gilvum_PYR-GCK TERGGISLRVVADGAATSRPLRMMRIDEVIDLYATTDEAKAGRPA-----
TH_0498|M.thermoresistible__bu VSP-AVNLTIVADGPATSRPLKLLGIADLVPLHATLDEALAAVAT-----
Mvan_2530|M.vanbaalenii_PYR-1 CARSDVDWVIVG-SPAVRRLLAICDPDGELPLTDALSSALARLDRLAHGG
:. :*. . . * : : : . * :
Rv1904|M.tuberculosis_H37Rv --------------
Mb1939|M.bovis_AF2122/97 --------------
MMAR_2800|M.marinum_M --------------
MUL_2925|M.ulcerans_Agy99 --------------
MAV_3518|M.avium_104 --------------
Mflv_4938|M.gilvum_PYR-GCK --------------
TH_0498|M.thermoresistible__bu --------------
Mvan_2530|M.vanbaalenii_PYR-1 QHLVWTAKSGWHAR